We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Samuel Sullivan/Sandbox 1
From Proteopedia
< User:Samuel Sullivan(Difference between revisions)
| Line 5: | Line 5: | ||
[[Image:ABCG2_without_Fab.png|400 px|right|thumb|Figure 1|]] | [[Image:ABCG2_without_Fab.png|400 px|right|thumb|Figure 1|]] | ||
==General Structure== | ==General Structure== | ||
| - | <scene name='83/837247/Fab_binding_site/1'>Fab binding site</scene> | ||
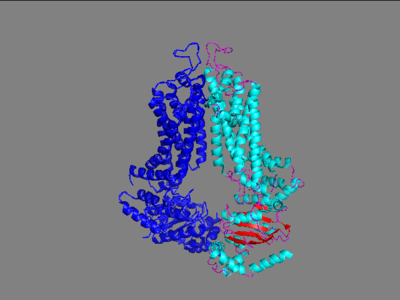
The ABCG2 protein is comprised of a homodimer which each have two specific domains: one spanning the cell membrane and one involved with nucleotide binding. | The ABCG2 protein is comprised of a homodimer which each have two specific domains: one spanning the cell membrane and one involved with nucleotide binding. | ||
===Transmembrane Domains=== | ===Transmembrane Domains=== | ||
| - | In the transmembrane domain is located the leucine plug that separates the first site of binding from the second site of binding for the substrate. This is only opened when the protein is shifted from its interior facing to exterior facing formation by the transfer of a phosphate off of an ATP. This causes a shift and transfer to the second binding site which is open to the extracellular matrix. The Fab-FD3 antigen binding fragment was found to stabilize the protein in its inward facing conformation which allowed for high resolution images of the protein to be taken in this conformation. Without Fab attached, the conformation of the protein was constantly in flux, transporting substrates out of the cell constantly. Fab was found to only stabilize ABCG2 on the extracellular side. Three important interactions between Fab fragments and ABCG2 were important to binding stability and favorability. This binding occurred at the EL-3 of ABCG2 which is the helices that stretches the furthest from the cell membrane. These interactions were two disulfide bonds, one intramolecularly and the other intermolecularly, and an n-glycosylation site at residue Asn-596. | + | In the transmembrane domain is located the leucine plug that separates the first site of binding from the second site of binding for the substrate. This is only opened when the protein is shifted from its interior facing to exterior facing formation by the transfer of a phosphate off of an ATP. This causes a shift and transfer to the second binding site which is open to the extracellular matrix. The Fab-FD3 antigen binding fragment was found to stabilize the protein in its inward facing conformation which allowed for high resolution images of the protein to be taken in this conformation. Without Fab attached, the conformation of the protein was constantly in flux, transporting substrates out of the cell constantly. Fab was found to only stabilize ABCG2 on the extracellular side. Three important interactions between Fab fragments and ABCG2 were important to binding stability and favorability. This binding occurred at the EL-3 of ABCG2 which is the helices that stretches the furthest from the cell membrane. These interactions were two disulfide bonds, one intramolecularly and the other intermolecularly, and an n-glycosylation site at residue Asn-596. <scene name='83/837247/Fab_binding_site/1'>Fab binding site</scene> |
===Nucleotide Binding Domains=== | ===Nucleotide Binding Domains=== | ||
These two domains contain the active site of this transporter protein. The interest in this protein is in its involvement with mutidrug resistant cancer cells. This involvement is due to its active site's promiscuity as many xenobiotics have been found to transported to the outside of the cell by this transporter. | These two domains contain the active site of this transporter protein. The interest in this protein is in its involvement with mutidrug resistant cancer cells. This involvement is due to its active site's promiscuity as many xenobiotics have been found to transported to the outside of the cell by this transporter. | ||
Current revision
ABCG2 Transporter Protein
| |||||||||||
References
- ↑ Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NMI, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B, Buschauer A, Stahlberg H, Altmann KH, Locher KP. Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol. 2018 Apr;25(4):333-340. doi: 10.1038/s41594-018-0049-1. Epub, 2018 Apr 2. PMID:29610494 doi:http://dx.doi.org/10.1038/s41594-018-0049-1
- ↑ Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature. 2018 Nov;563(7731):426-430. doi: 10.1038/s41586-018-0680-3. Epub 2018 Nov, 7. PMID:30405239 doi:http://dx.doi.org/10.1038/s41586-018-0680-3
- ↑ Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017 Jun 22;546(7659):504-509. doi: 10.1038/nature22345. Epub 2017 May, 29. PMID:28554189 doi:http://dx.doi.org/10.1038/nature22345
Student Contributors
Samuel Sullivan Jailyn Voyles Shelby Skaggs