We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Daniel Mulawa/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
== Structural highlights == | == Structural highlights == | ||
===Substrate Structure=== | ===Substrate Structure=== | ||
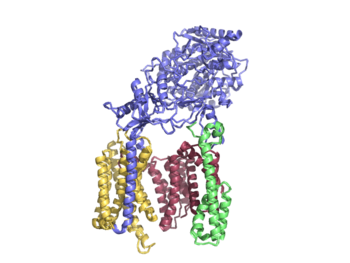
| - | + | Though gamma secretase has multiple substrates, the substrate of main concern is called Amyloid Precursor Protein (APP). APP is composed of an N-terminal loop, a transmembrane (TM) helix, and a C-terminal β-strand. The uses lateral diffusion as a mechanism of entry into the enzyme, and once in place, the TM helix is anchored by hydrogen bonds. In order to differentiate substrates, the β-strand is often the main point of identification for the enzyme. After this, the helix undergoes unwinding and the process of catalysis can begin. | |
| - | + | ||
TM helix anchored by hydrogen bonds | TM helix anchored by hydrogen bonds | ||
Unwinding model | Unwinding model | ||
| - | + | used for substrate recognition | |
===Lid Complex=== | ===Lid Complex=== | ||
| - | + | Lid is the first point of entry and recognition for the substrate. | |
===Active Site=== | ===Active Site=== | ||
Revision as of 20:36, 24 March 2020
Gamma Secretase
=Human Gamma Secretase=
| |||||||||||
References
1.Bai X, Yan C, Guanghui Yang, et al. 2015. An atomic structure of human γ-secretase. Nature. 525:212-217. 2.Carroll CM, and Li YM. 2016. Physiological and pathological roles of the γ-secretase complex. Brain research bulletin. 126:199-206. 3.Yang G, Zhou R, Shi Y. 2017. Cryo-EM structures of human γ-secretase. Current Opinion in Structural Biology. 46:55–64. 4.Yang G, Zhou R, Zhou Q, et al. 2019. Structural basis of Notch recognition by human γ-secretase. Nature. 565: 192-197. 5.Zhou R, Yang G, Guo X, et al. 2019. Recognition of the amyloid precursor protein by human γ-secretase. Science. 363:1-8.
Student Contributors
Layla Wisser Daniel Mulawa