This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Johnson's Monday Lab Sandbox for Insulin Receptor
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
===Conformational Changes=== | ===Conformational Changes=== | ||
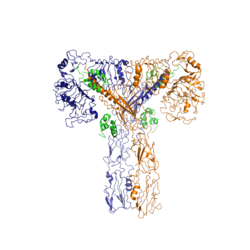
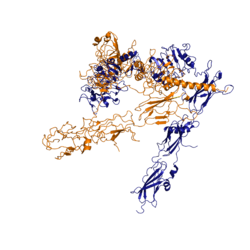
[[Image:image 6.png|thumb|left|250px|Figure 3: Conformational change of insulin receptor protomer from inactive (blue) to active (orange) form upon insulin binding. Inactive state PDB: 4zxb. Active state PDB: 6sof]] | [[Image:image 6.png|thumb|left|250px|Figure 3: Conformational change of insulin receptor protomer from inactive (blue) to active (orange) form upon insulin binding. Inactive state PDB: 4zxb. Active state PDB: 6sof]] | ||
| - | The conformational change between the inverted, inactive <scene name='83/839263/V_shape/3'>"V" shape</scene> and the active <scene name='83/839263/T-shape/4'>"T" shape</scene> of the insulin receptor is induced by insulin binding. When an insulin molecule binds to site 1 of the alpha subunit, the respective protomer is recruited and a slight inward movement of the <scene name='83/839263/Fniii_domains/1'>Fibronectin type III domains</scene> of the beta subunit is initiated. This is accomplished by the formation of several [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridges], specifically between <scene name='83/839263/Salt_bridges/1'>Arg498 and Asp499 of the FnIII-1 and Lys703, Glu706, and Asp707 of the alpha-CT</scene> <ref name="Uchikawa" />. Binding of insulin to both protomers establishes a full activation of the insulin receptor. This activation is demonstrated through the inward movement of both protomers. This motion has been referred to as a "hinge" motion <ref name="Uchikawa" /> as both protomers "swing" in towards one another. Figure 3 depicts the conformational change and "hinge motion" between the inactive and active forms of an insulin receptor protomer. | + | The conformational change between the inverted, inactive <scene name='83/839263/V_shape/3'>"V" shape</scene> and the active <scene name='83/839263/T-shape/4'>"T" shape</scene> of the insulin receptor is induced by insulin binding. When an insulin molecule binds to site 1 of the alpha subunit, the respective protomer is recruited and a slight inward movement of the <scene name='83/839263/Fniii_domains/1'>Fibronectin type III domains</scene> of the beta subunit is initiated. This is accomplished by the formation of several [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridges], specifically between <scene name='83/839263/Salt_bridges/1'>Arg498 and Asp499 of the FnIII-1 and Lys703, Glu706, and Asp707 of the alpha-CT</scene> <ref name="Uchikawa" />. Binding of insulin to both protomers establishes a full activation of the insulin receptor. This activation is demonstrated through the inward movement of both protomers. This motion has been referred to as a "hinge" motion <ref name="Uchikawa" /> as both protomers "swing" in towards one another. Figure 3 depicts the conformational change and "hinge motion" between the inactive and active forms of an insulin receptor protomer. Upon insulin binding, the beta subunits of the inactive form, shown in blue, are "swung" inward to the active form, shown in orange. |
As the fibronectin type III domains of the beta subunit swing inward, the alpha subunits also undergo a conformational change upon insulin binding. As insulin binds to site 1, the leucine-rich region of one protomer interacts with the ''alpha''-CT and the FNIII-1 domains of the other protomer to form a binding site. These interactions are referred to as the <scene name='83/839263/Tripartite_interface/2'>tripartite interface</scene> <ref name="Uchikawa" />. In order for the tripartite interface to form, the alpha subunits of each protomer must undergo a "folding" motion. | As the fibronectin type III domains of the beta subunit swing inward, the alpha subunits also undergo a conformational change upon insulin binding. As insulin binds to site 1, the leucine-rich region of one protomer interacts with the ''alpha''-CT and the FNIII-1 domains of the other protomer to form a binding site. These interactions are referred to as the <scene name='83/839263/Tripartite_interface/2'>tripartite interface</scene> <ref name="Uchikawa" />. In order for the tripartite interface to form, the alpha subunits of each protomer must undergo a "folding" motion. | ||
| Line 33: | Line 33: | ||
There are a multitude of hypotheses which discuss the reasons for the development of type II diabetes <ref name="Boucher" /> <ref name="Franks" />. Historically, the chronic condition has been closely associated with high caloric intake and sedentary lifestyles. However, recent studies, which have evaluated the relationships between genetics and environmental factors in the development of T2D, have shown that T2D is not uniform among the population and is more complicated than simply diet and exercise <ref name="Franks" />. A variety of factors may play a role in risk for T2D including gestational environment, [http://en.wikipedia.org/wiki/Human_microbiome microbiome], genetics, diet, and energy expenditure <ref name="Franks" />. Furthermore, the possible genetic or environmental factors which contribute to the development of T2D do not follow the same biochemical pathway to initiate insulin resistance. The establishment of insulin resistance is complex at both the macroscopic and molecular levels. | There are a multitude of hypotheses which discuss the reasons for the development of type II diabetes <ref name="Boucher" /> <ref name="Franks" />. Historically, the chronic condition has been closely associated with high caloric intake and sedentary lifestyles. However, recent studies, which have evaluated the relationships between genetics and environmental factors in the development of T2D, have shown that T2D is not uniform among the population and is more complicated than simply diet and exercise <ref name="Franks" />. A variety of factors may play a role in risk for T2D including gestational environment, [http://en.wikipedia.org/wiki/Human_microbiome microbiome], genetics, diet, and energy expenditure <ref name="Franks" />. Furthermore, the possible genetic or environmental factors which contribute to the development of T2D do not follow the same biochemical pathway to initiate insulin resistance. The establishment of insulin resistance is complex at both the macroscopic and molecular levels. | ||
| - | Current molecular explanations for insulin resistance include lipotoxicity, inflammation, reactive oxygen species, endoplasmic reticulum stress, and hyperglycemia <ref name="Boucher" />. Under normal conditions, the signal from the insulin receptor is transduced to the insulin receptor substrate (IRS-1) upon phosphorylation of tyrosine residues on IRS-1. In some T2D cases, serine residues of IRS-1 are phosphorylated instead of tyrosine residues and the signal from the insulin receptor is no longer properly conducted intracellularly. This | + | Current molecular explanations for insulin resistance include lipotoxicity, inflammation, reactive oxygen species, endoplasmic reticulum stress, and hyperglycemia <ref name="Boucher" />. Under normal conditions, the signal from the insulin receptor is transduced to the [http://proteopedia.org/wiki/index.php/5u1m insulin receptor substrate (IRS-1)] upon phosphorylation of tyrosine residues on IRS-1. From there, IRS-1 initiates a series of cascades, one of which is glucose transport by Glut4. In some T2D cases, serine residues of IRS-1 are phosphorylated instead of tyrosine residues and the signal from the insulin receptor is no longer properly conducted intracellularly. As a result, the glucose transport pathway is not stimulated, glucose is not moved into the cell, and blood glucose concentrations remain high <ref name="Boucher" />. This occurrence can be explained by a multitude of cellular stresses, namely lipotoxicity and the presence of reactive oxygen species (ROS). |
Reactive oxygen species are produced as a by-product of oxidative phosphorylation in the mitochondria. Increasing concentrations of ROS activate stress kinases which phosphorylate the serine residues of IRS-1 rather than the tyrosine residues <ref name="Boucher" />. Increased lipid content in cellular environments has also been shown to alter the phosphorylation of IRS-1. Diacylglycerol (DAG) is an [http://en.wikipedia.org/wiki/Intramyocellular_lipids intramyocellular lipid]. Increased concentrations of DAG in skeletal muscle activates the [http://en.wikipedia.org/wiki/Protein_kinase_C Protein kinase C] pathway, which induces phosphorylation of IRS-1 serine residues <ref name="Boucher" />. Increased circulation of fatty acids activates the same PKC pathway and also results in altered phosphorylation of IRS-1. Activation of PKC has also been demonstrated with increasing ceramide concentrations in the plasma membrane. As a result, the typical Protein kinase B (PKB) pathway of insulin signaling is inhibited <ref name="Boucher" />. | Reactive oxygen species are produced as a by-product of oxidative phosphorylation in the mitochondria. Increasing concentrations of ROS activate stress kinases which phosphorylate the serine residues of IRS-1 rather than the tyrosine residues <ref name="Boucher" />. Increased lipid content in cellular environments has also been shown to alter the phosphorylation of IRS-1. Diacylglycerol (DAG) is an [http://en.wikipedia.org/wiki/Intramyocellular_lipids intramyocellular lipid]. Increased concentrations of DAG in skeletal muscle activates the [http://en.wikipedia.org/wiki/Protein_kinase_C Protein kinase C] pathway, which induces phosphorylation of IRS-1 serine residues <ref name="Boucher" />. Increased circulation of fatty acids activates the same PKC pathway and also results in altered phosphorylation of IRS-1. Activation of PKC has also been demonstrated with increasing ceramide concentrations in the plasma membrane. As a result, the typical Protein kinase B (PKB) pathway of insulin signaling is inhibited <ref name="Boucher" />. | ||
Revision as of 14:07, 20 April 2020
Insulin Receptor
| |||||||||||