We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 896
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| + | |||
| + | ==Overview== | ||
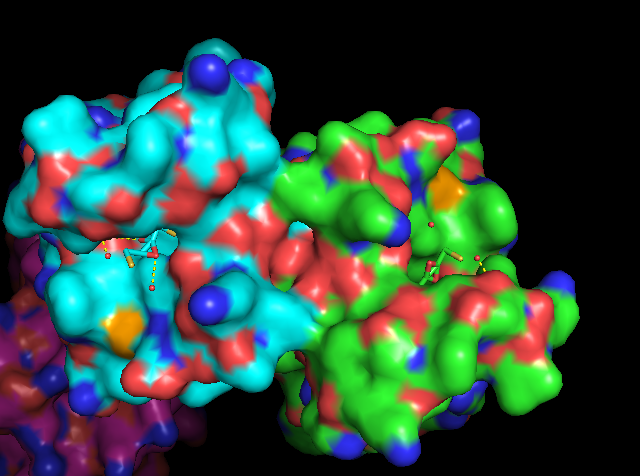
| + | Human bromodomain-containing protein 2 (BRD2) is a highly conserved and ubiquitously expressed protein involved in transcriptional regulation and recognition of post-translational histone modifications. BRD2 is a bromodomain and extra-terminal domain (BET) family protein, which are characterized by the presence of two adjacent bromodomains as well as an extra-terminal domain. Bromodomain and extra terminal domain (BET) family proteins are a group of related proteins involved in the specific recognition of acetylated lysine residues on chromatin and subsequent transcriptional activation [2]. As a BET protein, BRD2 follows this structural motif with its structure consisting of bromodomain 1 (BRD2-BD1) at the N-terminus, bromodomain 2 (BRD2-BD2), and an N-extra-terminal (NET) domain. Human BRD2 is encoded by the gene BRD2, which consists of 11 exons that cover more than 6 kb of genomic DNA [1]. This gene was formerly known as really interesting new gene 3 (RING3) but was later renamed to BRD2. | ||
== Function == | == Function == | ||
| + | Human BRD2 protein is a Serine-Threonine kinase found ubiquitously amongst the nuclear envelope of all cell types. The activity of BRD2 is increased during cell proliferation. BRD2 specifically recognizes an N-acetyl-lysine residue at position 12 of histone H4 via a homo-2-mer complex of BD1 domains. The BRD2-BD1 domain recognizes the H4 tail only when lysine-12 is acetylated. This recognition is mediated through the binding of the hypoacetylated side chain of lysine at position 8 of H4 with the interface between dimerized BRD2-BD1 domains [3]. BRD2 has also shown in vitro interaction with N-acetyl-lysine at position 5 of K12, but this interaction has not been shown to involve lysine-8 of H4 [3]. It is presumed that one of the functions of this recognition is to prevent deletion or erasure of post-translational histone markers during the mitotic cell cycle. The transcriptional regulation activity of BRD2 is also mediated through its positive regulation of E2F-dependent cell cycle progression (direct stimulation of E2F reporter activity) [1]. As E2F’s central function is to promote the synthesis of proteins needed for G1 to S transition, BRD2-BD1 is directly implicated in the regulation of the cell cycle. | ||
| + | |||
| + | [[Image:BRD2BD1SurfaceView.png]] | ||
[[Image:ProteopediaTest.png]] | [[Image:ProteopediaTest.png]] | ||
Revision as of 15:59, 28 April 2020
| This Sandbox is Reserved from Jan 13 through July 31, 2020 for use in the course Protein Structure in Drug Discovery taught by Karen C. Glass at the ACPHS, Colchester, United States. This reservation includes Sandbox Reserved 895 through Sandbox Reserved 901. |
To get started:
More help: Help:Editing |
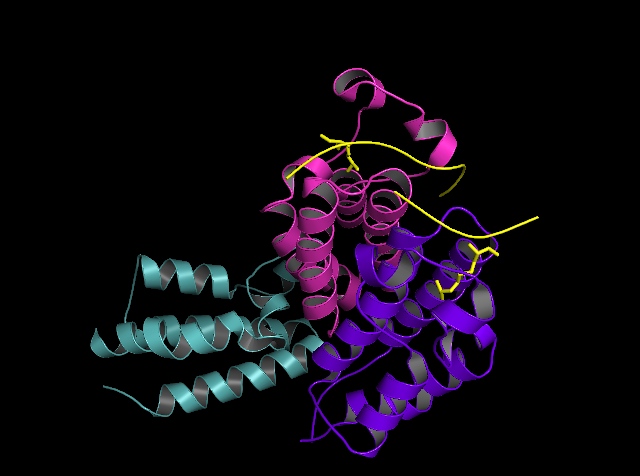
Structural and Functional Overview of Human BRD2 BD1 Bromodomain
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
|