User:Neal Hayhurst/RNA Polymerase II/Sandbox 1
From Proteopedia
| Line 42: | Line 42: | ||
| - | Source: https://www.pnas.org/content/94/1/151. | ||
| - | 1. <scene name='86/862225/Tfiid/5'>TFIID</scene> is highly conserved among eukaryotes. It recognizes and binds the TATA region of DNA. This is facilitated by a subunit named the TATA-binding protein (TBP). This subunit binding also causes major deformations in the helix which may be important for further binding of the PIC units. | ||
| - | + | 1. <scene name='86/862225/Tfiid/9'>TFIID</scene> is highly conserved among eukaryotes. It recognizes and binds the TATA region of DNA. This is facilitated by a subunit named the TATA-binding protein (TBP) that has antiparralel beta-sheet that provides a large surface for minor groove interactions. This subunit binding also causes major deformations in the helix which may be important for further binding of the PIC units by creating a more compact protein-DNA complex. As the complex begins to form the TBP-TATA complex remains unchanged. Its shape resembles that of a saddle sitting on the DNA. <ref name="txn"/> | |
| - | + | 2. <scene name='86/862225/Tfiib/3'>TFIIB</scene>> is the second to join the PIC. It is thought to be responsible for stabilizing the TBP/DNA complex and tethering the TFIID-DNA complex to RNAP I. It is also important in specifying the the TSS. In vitro studies have shown that accurate initiation can occur with only TFIID, TFIIB, and RNAP II suggesting that these two subunits serve to position RNAP. . Mutagenesis studies also suggest that it works as a spacer between TFIID and pol II. It may also function to ensure correct directionality. IF TBP binds the wrog end of TATA, TFIIB would have unfavorable interactions with TFB. <ref name="txn"/> | |
| - | + | 3. <scene name='86/862225/Tfiif/2'>TFIIF</scene> binds directly to RNAP II and forms a very stable complex. It then escorts RNAP II to the promoter TFIIF also increases specificity and efficiency of transcription. It also acts similarly to bacterial sigma factor by inhibiting and reversing RNAP II binding to nonpromoter sites. TFIIF is a hetero-dimer of 30 and 70 kDa. <ref name="txn"/> | |
| - | + | 4.<scene name='86/862225/Tfiie/6'>TFIEE</scene> is required to begin transcription even though RNAP II is bound to DNA before TFIIE binds. Once bound it recruits TFIIH. TfIIE is an α2β2 heterotetramer of 35 and 56 kDA. <ref name="txn"/> | |
| - | + | 5.<scene name='86/862225/Tfiih/3'>TFIIH</scene> supports catalytic activity such as DNA ATPase, DNA helicase, and a kinase that phosphorylates the CTD of RPB1.<ref name="txn"/>(F) Some of its subunits are also components of DNA repair machinery. It is the last TF to bind and completes the <scene name='82/824648/Pic/3'>PIC</scene>. | |
| + | |||
| + | 6.<scene name='86/862225/Tfiia/3'>TFIAA</scene> is a co-activator that helps regulate PIC assembly. It was initially thought to be essential for activity. It binds to and stabilizes the early complexes. It also neutralizes transcription repressors. The mechanism is unknown but it is thought to either increase TBP affinity for DNA or displace repressors. TFIIA binds to the N-terminal of TBP. It lies upstream, of TATA where it can interact with promoter and enhancer elements.<ref name="txn"/> | ||
Revision as of 01:05, 1 October 2020
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Young RA. RNA polymerase II. Annu Rev Biochem. 1991;60:689-715. doi: 10.1146/annurev.bi.60.070191.003353. PMID:1883205 doi:http://dx.doi.org/10.1146/annurev.bi.60.070191.003353
- ↑ 2.0 2.1 Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998 Oct 23;273(43):27757-60. doi: 10.1074/jbc.273.43.27757. PMID:9774381 doi:http://dx.doi.org/10.1074/jbc.273.43.27757
- ↑ 3.0 3.1 3.2 Sobennikova MV, Shematorova EK, Shpakovskii GV. [C-terminal domain (CTD) of the subunit Rpb1 of nuclear RNA polymerase II and its role in the transcription cycle]. Mol Biol (Mosk). 2007 May-Jun;41(3):433-49. PMID:17685222
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 RNA polymerase II transcription initiation: A structural view D. B. Nikolov, S. K. Burley Proceedings of the National Academy of Sciences Jan 1997, 94 (1) 15-22; DOI: 10.1073/pnas.94.1.15
- ↑ Hurwitz J. The discovery of RNA polymerase. J Biol Chem. 2005 Dec 30;280(52):42477-85. doi: 10.1074/jbc.X500006200. Epub 2005, Oct 17. PMID:16230341 doi:http://dx.doi.org/10.1074/jbc.X500006200
- ↑ doi: https://dx.doi.org/10.1038/nrm1796
- ↑ 7.0 7.1 7.2 7.3 Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
- ↑ He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 9.8 Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 10.8 10.9 Voet, D., Voet, J. G., & Pratt, C. W. (2013). Transcription and RNA Processing. In Fundamentals of biochemistry: life at the molecular level (pp. 933–942). Wiley.
- ↑ Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
- ↑ Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005 Jan 28;120(2):183-93. doi: 10.1016/j.cell.2004.11.045. PMID:15680325 doi:http://dx.doi.org/10.1016/j.cell.2004.11.045
Bushnell, D. A.; Westover, K. D.; Davis, R. E.; Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science. 2004, 303, 983-988
Brueckner, F. and Cramer, P. Structural Basis of Transcription Inhibition by -amanitin and Implications for RNA Polymerase II Translocation. Nature Structure and Molecular Biology. 2008, 15, 811-818.
Cramer, P.; Bushnell, D. A.; Kornberg, R. D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001, 292, 1863-1876
Evans, D. A.; Fitch, D. M.; Smith, T. E.; Cee, V. J. Application of Complex Aldol Reactions to the Total Synthesis of Phorboxazole B. J. Am. Chem. Soc. 2000, 122, 10033-10046.
Gnatt, A. L.; Cramer, P; Fu, J.; Bushnell, D. A.; and Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001, 292, 1876-1882 1i6h
Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
Shah, N. et. al. Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Molecular Cell. 2018, 69, 48-61.
Uzman, A.; Voet, D. Student companion Fundamentals of biochemistry: life at the molecular level, 4th ed., Donald Voet, Judith G. Voet, Charlotte W. Pratt; John Wiley & amp; Sons, 2012.
Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M. A.; Chong, J.; Hare, A. A.; Dervan, P. B.; DiMaio, F.; Leschziner, A. E.; Wang, D. Structural Basis for the Initiation of Eukaryotic Transcription-coupled DNA Repair. Nature. 2017. 551, 653-657 5vvr
Xin, L.; Bushnell, D. A.; and Kornburg, R. D. RNA Polymerase II Transcription: Structure and Mechanism. Biochemica et Biophysica Acta. 2013, 1829, 2-8.
Yan, C., Dodd, T., He, Y., Tainer, J. A., Tsutakawa, S. E., & Ivanov, I. (2019). Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural and Molecular Biology, 26(6), 397-406.
Alpha-aminitin chemical structure image courtesy of https://en.wikipedia.org/wiki/Alpha-Amanitin#/media/File:Alpha-amanitin_structure.png
Notes
From structural components:
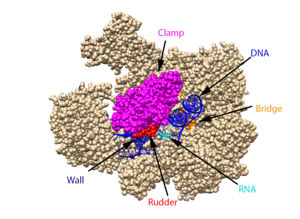
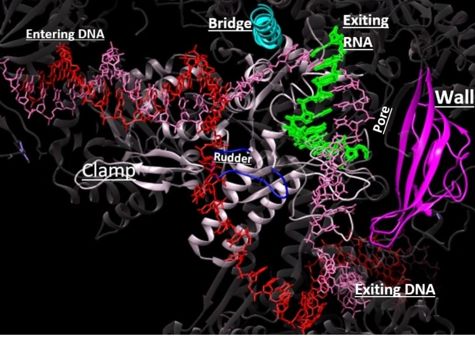
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019 and Fall of 2020. This page was reviewed by Dr. Wally Novak of Wabash College.