User:Neal Hayhurst/RNA Polymerase II/Sandbox 1
From Proteopedia
| Line 30: | Line 30: | ||
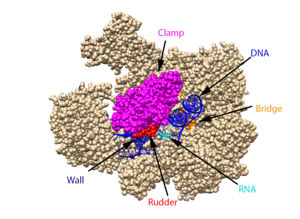
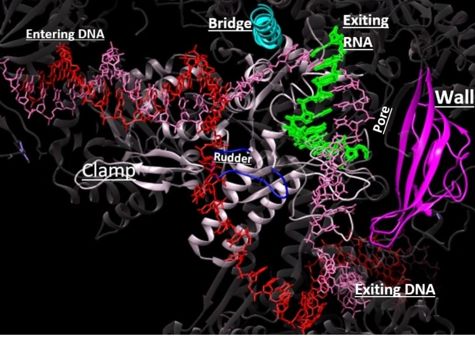
[[ Image:RnapII struct features.jpg|300px|left|thumb| The clamp (magenta), wall (navy blue), rudder (red), bridge (orange), RNA (light blue), and DNA (blue) are depicted. See below for PDB's and residue numbers.]] | [[ Image:RnapII struct features.jpg|300px|left|thumb| The clamp (magenta), wall (navy blue), rudder (red), bridge (orange), RNA (light blue), and DNA (blue) are depicted. See below for PDB's and residue numbers.]] | ||
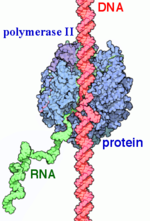
| - | DNA enters through the jaw, an opening in RNAP II. The <scene name='82/824648/Clamp/5'>clamp</scene> swings to trap the DNA in the cleft. The <scene name='82/824648/Wall/2'>wall</scene> sends the DNA template strand through the cleft in a turn of approximately 90°. Both the clamp and wall are parts of the Rpb2 subunit. <scene name='82/824648/Magnesium/3'>Magnesium</scene> is located within the active site and functions as the catalyst. NTPs travel through the funnel to RNAP II’s active site, where they are incorporated into the growing RNA strand. The Trigger loop helps RNAP II select a substrate ribonucleotide by ensuring that only NTPs that form a Watson-Crick base pair with the DNA template base enter the active site of RNAP II. The phosphorylation of the C-terminal domain (CTD), which is a part of the Rpb1 subunit, triggers the conversion of RNAP II’s initiation complex to RNAP II’s elongation complex. The <scene name='82/824648/Bridge/6'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each catalytic cycle. Further along in the process, the <scene name='82/824648/Rudder/3'>rudder</scene> separates the newly synthesized RNA strand from the DNA template strand, permitting the DNA double helix to reform as the DNA exits RNAP II. | + | DNA enters through the jaw, an opening in RNAP II. The <scene name='82/824648/Clamp/5'>clamp</scene> swings to trap the DNA in the cleft. The <scene name='82/824648/Wall/2'>wall</scene> sends the DNA template strand through the cleft in a turn of approximately 90°. Both the clamp and wall are parts of the Rpb2 subunit. <scene name='82/824648/Magnesium/3'>Magnesium</scene> is located within the active site and functions as the catalyst. NTPs travel through the funnel to RNAP II’s active site, where they are incorporated into the growing RNA strand. The Trigger loop helps RNAP II select a substrate ribonucleotide by ensuring that only NTPs that form a Watson-Crick base pair with the DNA template base enter the active site of RNAP II. The phosphorylation of the C-terminal domain (CTD), which is a part of the Rpb1 subunit, triggers the conversion of RNAP II’s initiation complex to <scene name='86/862212/Elongation_complex_pdb_2e2h/1'>RNAP II’s elongation complex</scene>. The <scene name='82/824648/Bridge/6'>bridge</scene> is an Rpb1 segment that translocates the DNA-RNA complex at the end of each catalytic cycle. Further along in the process, the <scene name='82/824648/Rudder/3'>rudder</scene> separates the newly synthesized RNA strand from the DNA template strand, permitting the DNA double helix to reform as the DNA exits RNAP II. |
| Line 76: | Line 76: | ||
===Elongation=== | ===Elongation=== | ||
| - | After a short transcript is synthesized during initiation, the process of elongation is stimulated by a conformational transition<ref name="VVP">Voet, D., Voet, J. G., & Pratt, C. W. (2013). Transcription and RNA Processing. In Fundamentals of biochemistry: life at the molecular level (pp. 933–942). Wiley.</ref>. To achieve the conformation conducive for elongation, the <scene name='86/862212/Tfiib_finger_domain/ | + | After a short transcript is synthesized during initiation, the process of elongation is stimulated by a conformational transition<ref name="VVP">Voet, D., Voet, J. G., & Pratt, C. W. (2013). Transcription and RNA Processing. In Fundamentals of biochemistry: life at the molecular level (pp. 933–942). Wiley.</ref>. To achieve the conformation conducive for elongation, the <scene name='86/862212/Tfiib_finger_domain/3'>TFIIB</scene> finger domain is displaced away from the active site to make room for the nascent RNA transcript and the CTD of the <scene name='86/862212/Rpb1_subunit/1'>Rpb1 subunit</scene> is phosphorylated<ref name="VVP"/><ref>DOI: 10.1073/pnas.140202297</ref>. Phosphorylation releases some initiation factors--some of these GTFs are left on the promoter region of the template to recruit another RNAPII<ref name="VVP"/>. <scene name='86/862212/Elongator_complex/1'>Elongator complex</scene> replaces the GTFs on the phosphorylated CTD of Rpb1 to accelerate transcription<ref name="VVP"/>. |
| - | DNA unwinds 3 nucleotides ahead of the active site (contained in Rpb1)<ref name="VVP"/>. Past the active site, the <scene name='86/862212/Wall_domain/ | + | DNA unwinds 3 nucleotides ahead of the active site (contained in Rpb1)<ref name="VVP"/>. Past the active site, the <scene name='86/862212/Wall_domain/1'>wall domain</scene> of Rpb2 redirects the template strand at about a 90o angle out of the cleft, orienting the template base in the active site towards the active site floor to align with the incoming NTP<ref name="VVP"/>. NTPs enter the active site on the floor side through a 12Å funnel called the pore, large enough for only one NTP at a time<ref name="VVP"/>. Once they have passed through the funnel, the NTPs enter <scene name='86/860989/Active_site_wn_test/1'>active site</scene> which is coordinated by three magnesium ions<ref name="Wang">DOI: 10.1016/j.cell.2006.11.023</ref>. |
| - | The <scene name='86/862212/Trigger_loop_and_bridge_helix/ | + | The <scene name='86/862212/Trigger_loop_and_bridge_helix/1'>trigger loop and bridge helix</scene> domains of Rpb1 are found adjacent to the active site and have been implicated in RNAPII NTP selectivity, catalysis, and translocation<ref name="Wang" /><ref name="Mishanina">DOI: 10.1073/pnas.1702383114</ref>. The trigger loop oscillates between positions near downstream DNA and near the active site<ref name="Wang" /><ref name="Mishanina" />. When a correctly paired NTP enters the A site, the trigger loop swings under the NTP, closing off the active site and allowing phosphodiester bond formation between the 3' end of the nascent RNA and the 5' end of the NTP<ref name="Wang" />. The trigger loop is stabilized by extensive interactions with the bridge helix, the NTP ribose, base, and phosphate groups, and other active site residues<ref name="Wang" /><ref name="Da">DOI:10.1021/ja210656k</ref>. The extensive trigger loop contacts slightly unwind and bend the bridge helix, a conformational change thought to play an important role in translocation<ref name="Wang" />. <scene name='86/862212/Trigger_loop/1'>HIS1085</scene> of the trigger loop has been proposed to catalyze phosphodiester bond formation by acting as a general acid<ref name="Wang" />. However, contradicting reports suggest that HIS1085 is not capable of acid catalysis, but rather catalyzes translocation as a positional catalyst<ref name="Mishanina" />. Release of a pyrophosphate group after bond formation destabilizes interactions with His1085, releasing the trigger loop from the active site and allowing movement of the DNA-RNA hybrid helix and entrance of a new NTP<ref name="Wang" /><ref name="Da" /><ref name="Mishanina" />. Becasue the trigger loop maintains the bridge helix conformation, its release allows the bridge helix to relax and such movement likely contributes to RNAPII translocation<ref name="Wang" />. |
====Kinetics==== | ====Kinetics==== | ||
Multiple kinetic models have been proposed with largely a consensus on some form of a Brownian ratchet mechanism involving the trigger loop and bridge helix in which the RNAPII has both forward and backward motion with a preference for forward translocation<ref>DOI: 10.1016/j.cell.2004.11.045</ref><ref name="Mishanina" />. | Multiple kinetic models have been proposed with largely a consensus on some form of a Brownian ratchet mechanism involving the trigger loop and bridge helix in which the RNAPII has both forward and backward motion with a preference for forward translocation<ref>DOI: 10.1016/j.cell.2004.11.045</ref><ref name="Mishanina" />. | ||
| - | RNAPII has nearly infinite processivity due to the Rpb2 <scene name='86/862212/Clamp/ | + | RNAPII has nearly infinite processivity due to the Rpb2 <scene name='86/862212/Clamp/1'>clamp</scene> subunit (β homolog) swinging down over DNA to trap it in the cleft<ref name="VVP" />. |
| Line 91: | Line 91: | ||
===Termination=== | ===Termination=== | ||
| - | The CTD has 26 heptapeptide repeats of the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (Schreieck et al, 2014). The phosphorylation of Tyr1 results in the recruitment of elongation factors and helps prevent premature recruitment of termination factors during transcription (Schreieck et al, 2014). The dephosphorylation of CTD ensures transition from elongation to termination. This dephosphorylation occurs at Tyr1 of the CTD and is catalyzed by the Glc7 subunit of the cleavage and polyadenylation factor (CPF) (Schreieck et al, 2014). Dephosphorylated Tyr1 results in the recruitment of pcf11. Pcf11 is able to bind to the phosphorylated Ser2 of CTD through its CTD-interacting domain (CID), resulting in the dissociation of the RNA pol II and the newly synthesized transcript from the DNA (Schreieck et al, 2014) (Zhang et al, 2005). Before the transcript is released, its 3’ end is polyadenylated. The DNA is brought back together at the other end of the transcription bubble, returning it to its original double-stranded form. | + | The CTD has 26 heptapeptide repeats of the consensus sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 (Schreieck et al, 2014). The phosphorylation of Tyr1 results in the recruitment of elongation factors and helps prevent premature recruitment of termination factors during transcription (Schreieck et al, 2014). The dephosphorylation of CTD ensures transition from elongation to termination. This dephosphorylation occurs at Tyr1 of the CTD and is catalyzed by the Glc7 subunit of the cleavage and polyadenylation factor (CPF) (Schreieck et al, 2014). Dephosphorylated Tyr1 results in the recruitment of pcf11. Pcf11 is able to bind to the phosphorylated Ser2 of CTD through its <scene name='86/862212/Pcf11_cid/1'>CTD-interacting domain (CID)</scene>, resulting in the dissociation of the RNA pol II and the newly synthesized transcript from the DNA (Schreieck et al, 2014) (Zhang et al, 2005). Before the transcript is released, its 3’ end is polyadenylated. The DNA is brought back together at the other end of the transcription bubble, returning it to its original double-stranded form. |
Revision as of 03:06, 1 October 2020
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Young RA. RNA polymerase II. Annu Rev Biochem. 1991;60:689-715. doi: 10.1146/annurev.bi.60.070191.003353. PMID:1883205 doi:http://dx.doi.org/10.1146/annurev.bi.60.070191.003353
- ↑ 2.0 2.1 Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998 Oct 23;273(43):27757-60. doi: 10.1074/jbc.273.43.27757. PMID:9774381 doi:http://dx.doi.org/10.1074/jbc.273.43.27757
- ↑ 3.0 3.1 3.2 Sobennikova MV, Shematorova EK, Shpakovskii GV. [C-terminal domain (CTD) of the subunit Rpb1 of nuclear RNA polymerase II and its role in the transcription cycle]. Mol Biol (Mosk). 2007 May-Jun;41(3):433-49. PMID:17685222
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 RNA polymerase II transcription initiation: A structural view D. B. Nikolov, S. K. Burley Proceedings of the National Academy of Sciences Jan 1997, 94 (1) 15-22; DOI: 10.1073/pnas.94.1.15
- ↑ Hurwitz J. The discovery of RNA polymerase. J Biol Chem. 2005 Dec 30;280(52):42477-85. doi: 10.1074/jbc.X500006200. Epub 2005, Oct 17. PMID:16230341 doi:http://dx.doi.org/10.1074/jbc.X500006200
- ↑ doi: https://dx.doi.org/10.1038/nrm1796
- ↑ 7.0 7.1 7.2 7.3 Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
- ↑ 8.0 8.1 Xin, L.; Bushnell, D. A.; and Kornburg, R. D. RNA Polymerase II Transcription: Structure and Mechanism. Biochemica et Biophysica Acta. 2013, 1829, 2-8.
- ↑ Chang-Hui Shen; Diagnostic Molecular Biology, 2019
- ↑ He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 11.8 Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
- ↑ 12.0 12.1 Eick, D, Geyer, M.The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chemical Reviews, 2013, 113 (11), 8456-8490 DOI: 10.1021/cr400071f
- ↑ Wang W, Carey M, Gralla JD. Polymerase II Promoter Activation: Closed Complex Formation and ATP-Driven Start Site Opening. Science. 1992;255:450–453.
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 14.6 14.7 14.8 14.9 Voet, D., Voet, J. G., & Pratt, C. W. (2013). Transcription and RNA Processing. In Fundamentals of biochemistry: life at the molecular level (pp. 933–942). Wiley.
- ↑ Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
- ↑ Svetlov, V., & Nudler, E. (2013). Basic mechanism of transcription by RNA polymerase II. Biochimica et biophysica acta, 1829(1), 20–28. https://doi.org/10.1016/j.bbagrm.2012.08.009
- ↑ Castano E, Gross P, Wang Z, Roeder RG, Oelgeschlager T. The C-terminal domain-phosphorylated IIO form of RNA polymerase II is associated with the transcription repressor NC2 (Dr1/DRAP1) and is required for transcription activation in human nuclear extracts. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7184-9. doi: 10.1073/pnas.140202297. PMID:10852970 doi:http://dx.doi.org/10.1073/pnas.140202297
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 18.6 18.7 18.8 Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006 Dec 1;127(5):941-54. PMID:17129781 doi:http://dx.doi.org/10.1016/j.cell.2006.11.023
- ↑ 19.0 19.1 19.2 19.3 19.4 Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci U S A. 2017 Jun 27;114(26):E5103-E5112. doi:, 10.1073/pnas.1702383114. Epub 2017 Jun 12. PMID:28607053 doi:http://dx.doi.org/10.1073/pnas.1702383114
- ↑ 20.0 20.1 Da LT, Wang D, Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J Am Chem Soc. 2012 Feb 1;134(4):2399-406. doi: 10.1021/ja210656k. Epub 2012 Jan , 24. PMID:22206270 doi:http://dx.doi.org/10.1021/ja210656k
- ↑ Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005 Jan 28;120(2):183-93. doi: 10.1016/j.cell.2004.11.045. PMID:15680325 doi:http://dx.doi.org/10.1016/j.cell.2004.11.045
Bushnell, D. A.; Westover, K. D.; Davis, R. E.; Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science. 2004, 303, 983-988
Brueckner, F. and Cramer, P. Structural Basis of Transcription Inhibition by -amanitin and Implications for RNA Polymerase II Translocation. Nature Structure and Molecular Biology. 2008, 15, 811-818.
Cramer, P.; Bushnell, D. A.; Kornberg, R. D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001, 292, 1863-1876
Evans, D. A.; Fitch, D. M.; Smith, T. E.; Cee, V. J. Application of Complex Aldol Reactions to the Total Synthesis of Phorboxazole B. J. Am. Chem. Soc. 2000, 122, 10033-10046.
Gnatt, A. L.; Cramer, P; Fu, J.; Bushnell, D. A.; and Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001, 292, 1876-1882 1i6h
Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
Shah, N. et. al. Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Molecular Cell. 2018, 69, 48-61.
Uzman, A.; Voet, D. Student companion Fundamentals of biochemistry: life at the molecular level, 4th ed., Donald Voet, Judith G. Voet, Charlotte W. Pratt; John Wiley & amp; Sons, 2012.
Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M. A.; Chong, J.; Hare, A. A.; Dervan, P. B.; DiMaio, F.; Leschziner, A. E.; Wang, D. Structural Basis for the Initiation of Eukaryotic Transcription-coupled DNA Repair. Nature. 2017. 551, 653-657 5vvr
Xin, L.; Bushnell, D. A.; and Kornburg, R. D. RNA Polymerase II Transcription: Structure and Mechanism. Biochemica et Biophysica Acta. 2013, 1829, 2-8.
Yan, C., Dodd, T., He, Y., Tainer, J. A., Tsutakawa, S. E., & Ivanov, I. (2019). Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural and Molecular Biology, 26(6), 397-406.
Eick, D, Geyer, M.The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chemical Reviews, 2013, 113 (11), 8456-8490 DOI: 10.1021/cr400071f
Chang-Hui Shen; Diagnostic Molecular Biology, 2019
Alpha-aminitin chemical structure image courtesy of https://en.wikipedia.org/wiki/Alpha-Amanitin#/media/File:Alpha-amanitin_structure.png
Notes
From structural components:
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019 and Fall of 2020. This page was reviewed by Dr. Wally Novak of Wabash College.