We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1639
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
== Other important features == | == Other important features == | ||
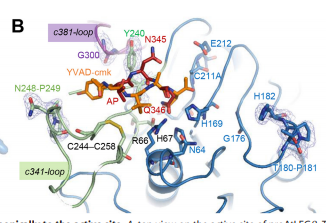
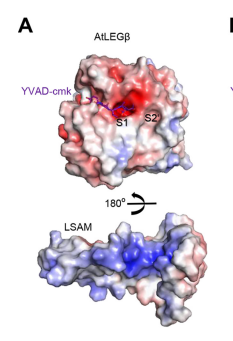
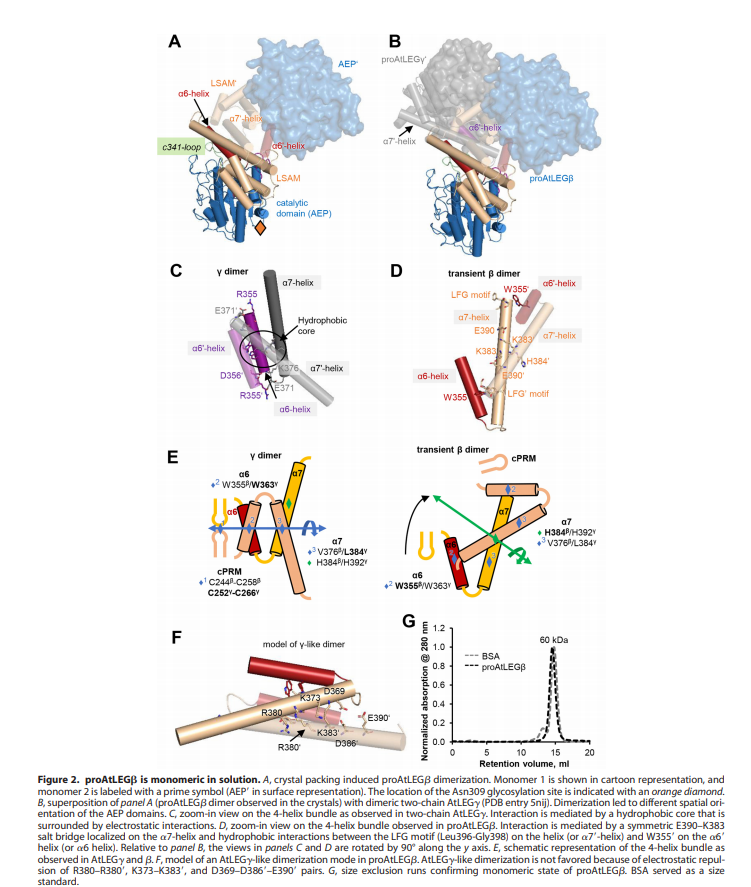
| - | An important feature is that proAtLEGb forms atypical dimers in the crustal and is monomeric in solutions. When compared to another form of our protein AtLEGgy findings suggest that the observed beta-dimer is weak and probably only transient in solution. [[Image | + | An important feature is that proAtLEGb forms atypical dimers in the crustal and is monomeric in solutions. When compared to another form of our protein AtLEGgy findings suggest that the observed beta-dimer is weak and probably only transient in solution. [[Image:Monomeric.png]] |
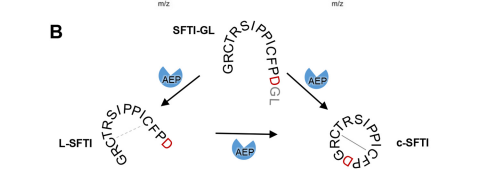
Another important feature is the cyclization of the peptides after cleavage. The cyclization is important for the cell stays programmed and is the same every time. [[Image:Cyclization.png]] | Another important feature is the cyclization of the peptides after cleavage. The cyclization is important for the cell stays programmed and is the same every time. [[Image:Cyclization.png]] | ||
Revision as of 05:32, 8 December 2020
| This Sandbox is Reserved from 09/18/2020 through 03/20/2021 for use in CHEM 351 Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, IA. This reservation includes Sandbox Reserved 1628 through Sandbox Reserved 1642. |
To get started:
More help: Help:Editing |
Arabidopsis thaliana legumain isoform beta in zymogen state (6YSA)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
[1] PMID:32719006