|
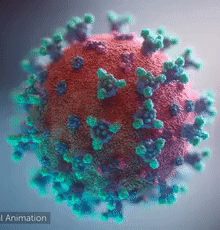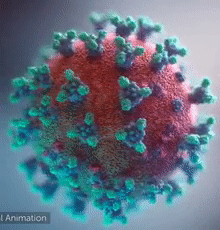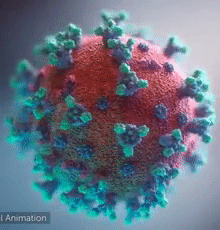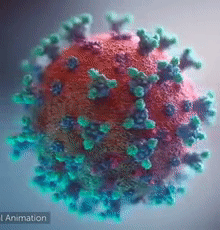 SARS-CoV-2 virus. The spikes, that adorn the virus surface, impart a corona like appearance (Fusion Animation).
A novel coronavirus SARS-CoV-2, detected in Wuhan, China in 2019, was found to cause severe respiratory illness named COVID-19[1].
There’s a new NCBI site on COVID-19/SARS-CoV-2. It will help bench scientists, bioinformaticians, clinicians & others connect with information they need to study SARS-CoV-2 and end the pandemic.
A video (by Elara) shows how the virus interacts with its human (host) cells, via its spike proteins[2][3][4][5][6][7], thus permitting the viral genome to enter its host & begin infection. The spike protein is induced for optimal binding when it is clipped by a protease (see animation on right side) and more details at SARS-CoV-2_protein_S_activation_by_furin. Subsequently, it undergoes a dramatic transformation to induce membrane fusion and infection, explained with a morph animation at SARS-CoV-2 spike protein fusion transformation.
Potential treatments for COVID-19
- 23-Mar-2020 A USA, French & UK study identified 69 drugs to test against the coronavirus[8]. As reported in the NY Times.
Videos helping to explain COVID-19
The Czech Republic took the uncommon step of making wearing of masks mandatory in public spaces, prompting a grassroots effort to make masks, with wonderful results
.
Fighting Coronavirus with Soap by PDB-101.
Modeling an epidemic (6-Mar-2020) We're not ready for the next epidemic by 3blue1brown.
COVID-19 outbreak shown via 3D animation (11-Feb 11-2020) by Sci Animations.
Bill Gates Ted Talk (3-Apr-2015) We're not ready for the next epidemic by Ted Talks.
George W. Bush urged us to prepare for future pandemics in 2005, at the NIH.
Jürgen Bosch Presentation (31-Mar-2020) Coronavirus: How it Ticks and What we are Doing to Stop it by Jürgen Bosch.
Useful sites on COVID-19
- What You Need to Know About the COVID-19 and HIV in English and Spanish from TheBody (The HIV/AIDS Resource).
- Coronavirus Evolved Naturally, and ‘Is Not a Laboratory Construct,’ in a study in Nature Med by Anderson and colleagues [10].
- A computer game, developed at the Inst for Protein Design (Univ Washington), is being used to try to find new lead compounds that might become drugs to treat COVID-19.
3D structural studies on SARS-CoV-2 virus
- A team of Chinese scientists determined, by Cryo-EM, the coronavirus spike receptor-binding domain complexed with its receptor ACE2 PDB-ID 6lzg (To be published).
- A team of US and Chinese scientists determined the crystal structure of 2019-nCoV spike receptor-binding domain bound with ACE2 6m0j
- Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 6w41
- Crystal Structure of the methyltransferase-stimulatory factor complex of NSP16 and NSP10 from SARS CoV-2 6w61 (To be published).
- Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP 6w6y (To be published).
- Structure of NSP10 - NSP16 Complex from SARS-CoV-2 6w75 (To be published).
- Crystal structure of SARS-CoV-2 nucleocapsid protein N-terminal RNA binding domain 6m3m (To be published).
- Crystal structure of Nsp9 RNA binding protein of SARS CoV-2 6w4b (To be published).
- Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2 6w4h (To be published).
- A Cryo-EM study by Zhou & colleagues on the structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2 gives insights to the molecular basis for coronavirus recognition and infection[12] 6m17.
- Crystal structure of RNA binding domain of nucleocapsid phosphoprotein from SARS coronavirus 2 6vyo (To be published).
- Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate 6w01 (To be published).
- Crystal structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in the complex with ADP ribose 6w02 (To be published).
- Crystal structure of 2019-nCoV chimeric receptor-binding domain complexed with its receptor human ACE2 6vw1
- Crystal structure of NSP15 Endoribonuclease from SARS CoV-2 6vww (To be published).
- Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 6vxs (To be published).
- Crystal structure of the 2019-nCoV HR2 Domain 6lvn (To be published).
- Crystal structure of post fusion core of 2019-nCoV S2 subunit 6lxt.
- Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, from the Hilgenfeld lab[13], Apo Struture: PDB-ID 6y2e, and complexes with inhibitors: PDB-ID 6y2f and 6y2g.
- 3D Structure of RNA-dependent RNA polymerase from COVID-19, a major antiviral drug target from the Rao lab in Beijing[14].
- Crystal structure of the Mpro from COVID-19 and discovery of inhibitors in a study by scientists from Shanghai & Beijing[15], PDB-ID 6lu7.
- Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2 in a study by scientists from USA[16], PDB-ID 6w01.
- The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. A study by McLellan and colleagues in "Science" on the Cryo-EM structure of the COVID-19 spike protein. This structure should greatly aid in the rapid development and evaluation of medical countermeasures to address the ongoing public health crisis[3], PDB-ID 6vsb.
SARS-CoV-2 virus proteins
The genome of the SARS-CoV-2 virus codes for 28 proteins:
Out of those, 19 have already been characterized structurally.
Details of the 3D structure & function of the key proteins & RNA inside the virus can be seen in the NY Times Bad News Wrapped in Protein: Inside the Coronavirus Genome.
References
- ↑ Naming the coronavirus disease (COVID-19) and the virus that causes it
- ↑ Pillay TS. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J Clin Pathol. 2020 Jul;73(7):366-369. doi: 10.1136/jclinpath-2020-206658. Epub, 2020 May 6. PMID:32376714 doi:http://dx.doi.org/10.1136/jclinpath-2020-206658
- ↑ 3.0 3.1 Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Feb 19. pii: science.abb2507. doi: 10.1126/science.abb2507. PMID:32075877 doi:http://dx.doi.org/10.1126/science.abb2507
- ↑ Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Mar 6. pii: S0092-8674(20)30262-2. doi: 10.1016/j.cell.2020.02.058. PMID:32155444 doi:http://dx.doi.org/10.1016/j.cell.2020.02.058
- ↑ Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020 Jul 9. pii: 10.1038/s41594-020-0468-7. doi:, 10.1038/s41594-020-0468-7. PMID:32647346 doi:http://dx.doi.org/10.1038/s41594-020-0468-7
- ↑ Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002 Oct;3(10):753-66. doi: 10.1038/nrm934. PMID:12360192 doi:http://dx.doi.org/10.1038/nrm934
- ↑ Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020 Apr 7. pii: S0092-8674(20)30338-X. doi: 10.1016/j.cell.2020.03.045. PMID:32275855 doi:http://dx.doi.org/10.1016/j.cell.2020.03.045
- ↑ Gordon, et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing: bioRxiv (online) 2020 http://doi.org/10.1101/2020.03.22.002386
- ↑ Ruffell D. Coronavirus SARS-CoV-2: filtering fact from fiction in the infodemic: Q&A with virologist Professor Urs Greber. FEBS Lett. 2020 Apr 4. doi: 10.1002/1873-3468.13784. PMID:32246722 doi:http://dx.doi.org/10.1002/1873-3468.13784
- ↑ Andersen, et al. The proximal origin of SARS-CoV-2: Nature Med (in press) 2020 http://dx.doi.org/10.1038/s41591-020-0820-9]
- ↑ Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Mar 6. pii: S0092-8674(20)30262-2. doi: 10.1016/j.cell.2020.02.058. PMID:32155444 doi:http://dx.doi.org/10.1016/j.cell.2020.02.058
- ↑ Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 4. pii: science.abb2762. doi: 10.1126/science.abb2762. PMID:32132184 doi:http://dx.doi.org/10.1126/science.abb2762
- ↑ Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020 Mar 20. pii: science.abb3405. doi: 10.1126/science.abb3405. PMID:32198291 doi:http://dx.doi.org/10.1126/science.abb3405
- ↑ Gao, et al. Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug target: bioRxiv (online) 2020 http://doi.org/10.1101/2020.03.16.993386
- ↑ Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020 Apr 9. pii: 10.1038/s41586-020-2223-y. doi:, 10.1038/s41586-020-2223-y. PMID:32272481 doi:http://dx.doi.org/10.1038/s41586-020-2223-y
- ↑ Kim, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2: bioRxiv (online) 2020 http://doi.org/10.1101/2020.03.02.968388
|

