User:Abbey Wells/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
Mechanistically, this reaction involves a molecular oxygen, water molecule, and the transport of electrons down an [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain] consisting of cytochrome b5 reductase, cytochrome b5, and NADH to the irons ions within SCD1 which then through a series of redox reactions introduces a double bond between the 9th and 10th carbons of Stearoyl-CoA forming oleic acid. | Mechanistically, this reaction involves a molecular oxygen, water molecule, and the transport of electrons down an [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain] consisting of cytochrome b5 reductase, cytochrome b5, and NADH to the irons ions within SCD1 which then through a series of redox reactions introduces a double bond between the 9th and 10th carbons of Stearoyl-CoA forming oleic acid. | ||
| - | == | + | == Structure == |
| - | == Binding of Substrate == | + | === Binding of Substrate === |
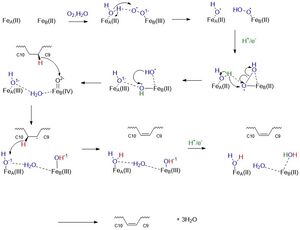
Stearoyl-CoA is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty acyl-CoA. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and many atoms of Nitrogen, oxygen, and sulfur. The fatty acid tail is a 17-carbon chain which reaches into the interior of the protein. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached via <scene name='87/877602/Hydrophillic_top/2'>Hydrophilic Top</scene> groups that include R151, D152, and K185. The rest of the exterior of the substrate is attached via <scene name='87/877602/Hydrophillic_bottom_labeled/2'>Hydrophilic Bottom</scene> groups that include R184, N144, and N71. The fatty acid chain dives into the interior of the enzyme. The acyl chain is enclosed in a tunnel (24 ang) buried in the cytosolic domain. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction. The chain is kinked at carbon 9-carbon 10 where the double bond is generated. The <scene name='87/877602/Full_kink/2'>Kink</scene> is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink/2'>W149, T257, Q143</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149. These residues are directly below the kink and will be hydrolyzed when the substrate is ready to be released. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/Trp258/2'>Trp258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. | Stearoyl-CoA is the substrate that binds to the enzyme, SCD1. The binding of the substrate is stabilized by specific residues on the exterior and interior of the protein. Stearoyl-CoA is a long-chain fatty acyl-CoA. The head group of the substrate is composed of an adenine, ribose, phosphate groups, and many atoms of Nitrogen, oxygen, and sulfur. The fatty acid tail is a 17-carbon chain which reaches into the interior of the protein. The head of stearoyl-CoA is attached to the exterior of the protein by polar residues. The adenine, ribose, and phosphate are attached via <scene name='87/877602/Hydrophillic_top/2'>Hydrophilic Top</scene> groups that include R151, D152, and K185. The rest of the exterior of the substrate is attached via <scene name='87/877602/Hydrophillic_bottom_labeled/2'>Hydrophilic Bottom</scene> groups that include R184, N144, and N71. The fatty acid chain dives into the interior of the enzyme. The acyl chain is enclosed in a tunnel (24 ang) buried in the cytosolic domain. The geometry of the tunnel and formation of bound acyl chain are the structural basis for the stereospecificity of the desaturation reaction. The chain is kinked at carbon 9-carbon 10 where the double bond is generated. The <scene name='87/877602/Full_kink/2'>Kink</scene> is induced through the interactions of four conserved residues. Three out of four of these residues are not bound to the chain, but are hydrogen bonded to each other: <scene name='87/877602/Kink/2'>W149, T257, Q143</scene>. T257 is hydrogen bonded to Q143, and Q143 is hydrogen bonded to W149. These residues are directly below the kink and will be hydrolyzed when the substrate is ready to be released. The residue that is directly hydrogen bonded to the chain is <scene name='87/877602/Trp258/2'>Trp258</scene>. This residue is highly conserved and stabilizes the chain so it will be in the correct orientation in the active site. | ||
| - | == Active Site == | + | === Active Site === |
Two structures of SCD known. One structure shows the substrate, water molecule, and zinc (4YMK). The second structure shows the product and iron (6WF2). For the pictures below, the structure is shown with two zinc ions in order to demonstrate how is <scene name='87/877627/Zn_and_water/1'>water</scene> coordination with the ions. Water is used in the mechanism, which is why we chose to use the structure with zinc to explain the active site. | Two structures of SCD known. One structure shows the substrate, water molecule, and zinc (4YMK). The second structure shows the product and iron (6WF2). For the pictures below, the structure is shown with two zinc ions in order to demonstrate how is <scene name='87/877627/Zn_and_water/1'>water</scene> coordination with the ions. Water is used in the mechanism, which is why we chose to use the structure with zinc to explain the active site. | ||
The two <scene name='87/877627/Just_zn/1'>zinc</scene>ions are 6.4 Angstroms apart. The ions sit above the kink in the active site. They are stabilized by the <scene name='87/877627/His_box2/1'>his box</scene>. The ion closest to C9 is 5.2 angstroms from it. This ion interacts with 4 histidines, H156, H265, H294, H298, and one water molecule. The ion closest to C10 is 4.7 angstroms away from it. This ion interacts with 5 histidines, H116, H121, H153, H157, and H297. These 9 total Histidine residues form a <scene name='87/877627/His_box2/1'>his box</scene>.The his box is used to stabilize the ions into the active site, forming a prosthetic group. The his box is highly conserved among the other isoforms of SCD. Other residues around the his box are used to hydrogen bond to the histidines to stabilize them. These residues include: <scene name='87/877627/E161/1'>E161,</scene> | The two <scene name='87/877627/Just_zn/1'>zinc</scene>ions are 6.4 Angstroms apart. The ions sit above the kink in the active site. They are stabilized by the <scene name='87/877627/His_box2/1'>his box</scene>. The ion closest to C9 is 5.2 angstroms from it. This ion interacts with 4 histidines, H156, H265, H294, H298, and one water molecule. The ion closest to C10 is 4.7 angstroms away from it. This ion interacts with 5 histidines, H116, H121, H153, H157, and H297. These 9 total Histidine residues form a <scene name='87/877627/His_box2/1'>his box</scene>.The his box is used to stabilize the ions into the active site, forming a prosthetic group. The his box is highly conserved among the other isoforms of SCD. Other residues around the his box are used to hydrogen bond to the histidines to stabilize them. These residues include: <scene name='87/877627/E161/1'>E161,</scene> | ||
Revision as of 16:33, 8 April 2021
Stearoyl-CoA Desaturase 1 from Mus musculus
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Abbey Wells
- Josey McKinley
- Anthony Durand