Journal:Molecular Cell:1
From Proteopedia
(Difference between revisions)

| Line 12: | Line 12: | ||
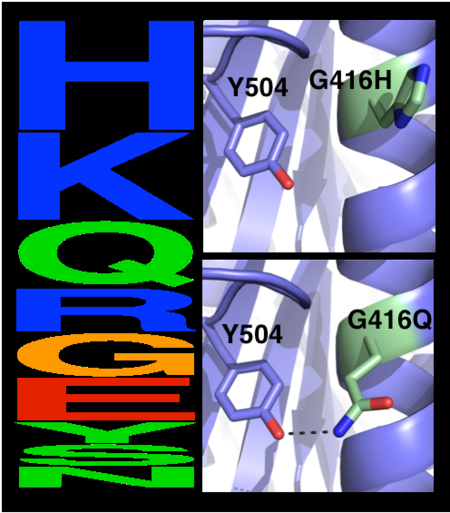
[[Image:MC1.png|left|450px|thumb|Eliminating potentially destabilizing mutations through homologous-sequence analysis and computational mutation scanning. Left: Sequence logo for hAChE position Gly416. The height of letters represents the respective amino acid’s frequency in an alignment of homologous AChE sequences. The evolutionarily ‘allowed’ sequence space (PSSM scores ≥0) at position 416 includes the 9 amino acids shown. Right: Structural models of mutations to the evolutionarily favored amino acid His, and to Gln, which is favored by Rosetta energy calculations. The His side chain is strained due to its proximity to the bulky Tyr504 aromatic ring, whereas the Gln side chain is relaxed and forms a favorable hydrogen bond with Tyr504 (dashed line)]] | [[Image:MC1.png|left|450px|thumb|Eliminating potentially destabilizing mutations through homologous-sequence analysis and computational mutation scanning. Left: Sequence logo for hAChE position Gly416. The height of letters represents the respective amino acid’s frequency in an alignment of homologous AChE sequences. The evolutionarily ‘allowed’ sequence space (PSSM scores ≥0) at position 416 includes the 9 amino acids shown. Right: Structural models of mutations to the evolutionarily favored amino acid His, and to Gln, which is favored by Rosetta energy calculations. The His side chain is strained due to its proximity to the bulky Tyr504 aromatic ring, whereas the Gln side chain is relaxed and forms a favorable hydrogen bond with Tyr504 (dashed line)]] | ||
{{Clear}} | {{Clear}} | ||
| - | Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), <font color='lime'><b> | + | Scenes highlight stabilizing effects of <scene name='72/728277/Cv/10'>selected mutations</scene> (in <font color='red'><b>red</b></font>), <font color='lime'><b>dAChE4 (PDB entry: [[5hq3]], green)</b></font> compared to <font color='cyan'><b>hAChE (PDB entry: [[4ey4]], cyan)</b></font>: |
*<scene name='72/728277/Cv/27'>Buried hydrogen bonds</scene>. | *<scene name='72/728277/Cv/27'>Buried hydrogen bonds</scene>. | ||
*<scene name='72/728277/Cv/28'>Surface polarity</scene>. | *<scene name='72/728277/Cv/28'>Surface polarity</scene>. | ||
Revision as of 14:34, 29 June 2021
| |||||||||||
- ↑ Goldenzweig A, Goldsmith M, Hill SE, Gertman O, Laurino P, Ashani Y, Dym O, Unger T, Albeck S, Prilusky J, Lieberman RL, Aharoni A, Silman I, Sussman JL, Tawfik DS, Fleishman SJ. Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability. Mol Cell. 2016 Jul 21;63(2):337-346. doi: 10.1016/j.molcel.2016.06.012. Epub 2016, Jul 14. PMID:27425410 doi:http://dx.doi.org/10.1016/j.molcel.2016.06.012
Proteopedia Page Contributors and Editors (what is this?)
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.