Journal:Acta Cryst D:S205979832100677X
From Proteopedia
(Difference between revisions)

| Line 11: | Line 11: | ||
*<scene name='88/886503/Cv/22'>The dimer formation of WcAG with 180ᴼ rotation</scene>. | *<scene name='88/886503/Cv/22'>The dimer formation of WcAG with 180ᴼ rotation</scene>. | ||
| - | ''Wc''AG formed a <scene name='88/886503/Cv/26'>homodimer, of which the N-terminal domain of one monomer orientated in proximity to the catalytic domain of another</scene>, creating the substrate-binding groove. The residues near the dimer interface are shown in dark blue and deep sky blue sticks, while the maltotriose and catalytic residues are represented by yellow and magenta, respectively | + | ''Wc''AG formed a <scene name='88/886503/Cv/26'>homodimer, of which the N-terminal domain of one monomer orientated in proximity to the catalytic domain of another</scene>, creating the substrate-binding groove. The residues near the dimer interface are shown in dark blue and deep sky blue sticks, while the maltotriose and catalytic residues are represented by yellow and magenta, respectively. |
Ligand binding sites: | Ligand binding sites: | ||
| Line 23: | Line 23: | ||
cyan/yellow, blue and red, respectively. The three catalytic residues are shown in salmon sticks. | cyan/yellow, blue and red, respectively. The three catalytic residues are shown in salmon sticks. | ||
| - | <scene name='88/886503/Cv2/10'>The active site of E374Q with bound maltotriose</scene>. | + | The active site of ''Wc''AG was naturally designed to fit perfectly with maltotriose. <scene name='88/886503/Cv2/10'>The active site of E374Q with bound maltotriose</scene>. E374/E374Q, D440, and D345/D345N are the catalytic residues (yellow) and other amino acid residues in/near the active site are shown in salmon. The asterisk and magenta colour present the residue from another subunit. The maltotriose is shown in cyan. White dashes represent hydrogen bonds. |
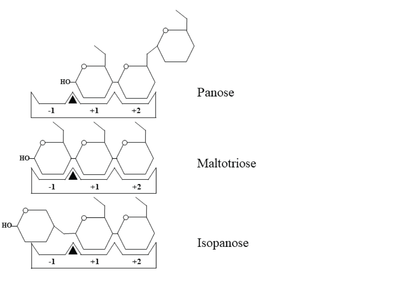
[[Image:Fig3BB.png|left|400px|thumb|Schematic representation of panose, maltotriose and isopanose binding within the active site of ''Wc''AG. The -1, +1, and +2 subsites are indicated as -1, +1, and +2, respectively. A black triangle represents the substrate cleavage site, located between subsites -1 and +1.]] | [[Image:Fig3BB.png|left|400px|thumb|Schematic representation of panose, maltotriose and isopanose binding within the active site of ''Wc''AG. The -1, +1, and +2 subsites are indicated as -1, +1, and +2, respectively. A black triangle represents the substrate cleavage site, located between subsites -1 and +1.]] | ||
Revision as of 12:45, 7 July 2021
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.