We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1692
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
== Other important features == | == Other important features == | ||
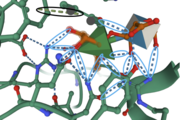
| - | The structure of FoRham1 consists of a <scene name='89/892735/Beta_propeller/4'>seven-bladed Beta-propellor domain</scene>, providing a favorable conformation for the substrate binding into the active site, which is located on the anterior of | + | The structure of FoRham1 consists of a <scene name='89/892735/Beta_propeller/4'>seven-bladed Beta-propellor domain</scene>, providing a favorable conformation for the substrate binding into the active site, which is located on the anterior side of the enzyme. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 04:53, 8 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
Structure and Function of FoRham1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Kondo T, Kichijo M, Maruta A, Nakaya M, Takenaka S, Arakawa T, Fushinobu S, Sakamoto T. Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family. J Biol Chem. 2021 Jul 22:101001. doi: 10.1016/j.jbc.2021.101001. PMID:34303708 doi:http://dx.doi.org/10.1016/j.jbc.2021.101001