We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1692
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Function of your protein == | == Function of your protein == | ||
| - | This enzyme, L-rhamnose- α-1,4-D-glucuronate lyase (FoRham1), is derived from the fungus ''Fusarium oxysporum'' and is a helpful tool for determining the structure and function of Gum Arabic (GA) to create potential agents to degrade GA more effectively. When the substrate GA is bound to FoRham1, the nonreducing ends of the glycosidic linkages are broken, releasing L-rhamnose (Rha) caps from GA | + | This enzyme, L-rhamnose- α-1,4-D-glucuronate lyase (FoRham1), is derived from the fungus ''Fusarium oxysporum'' and is a helpful tool for determining the structure and function of Gum Arabic (GA) to create potential agents to degrade GA more effectively. When the substrate GA is bound to FoRham1, the nonreducing ends of the glycosidic linkages are broken, releasing L-rhamnose (Rha) caps from GA <ref name="Kondo" />. Enzymes that can react with glycosidic linkages of certain carbohydrates can be useful in determining the structure, function, and mechanism of the carbohydrates, giving scientists the tools to manipulate their physical properties for further application to understand their breakdown. Determining this mechanism will give researchers more tools to understand how to degrade GA. Specifically, I focused on the mutant H105F, which has a PDB file of 7ESN. |
Shown here is the enzyme <scene name='89/892735/Protein_view_2/3'>with bound ligand </scene> using N->C coloring. | Shown here is the enzyme <scene name='89/892735/Protein_view_2/3'>with bound ligand </scene> using N->C coloring. | ||
== Biological relevance and broader implications == | == Biological relevance and broader implications == | ||
| - | Gum Arabic (GA) is a representative protein of the family of arabinogalactan proteins (AGPs) and is produced in acacia trees in response to stress conditions, such as drought or wounds. GA has a variety of applications within the industrial world, including the food, cosmetic, and pharmaceutical industries, acting specifically as an emulsion stabilizer, emulsifier, and thickener in pharmaceutical settings. However, the the detailed structure of GA has not determined because of the complex branching that occurs in the polysaccharide. Enzymes that can react with and eliminate glycosidic linkages of carbohydrates are useful for determining the structure and function of these carbohydrates, giving researchers the opportunity to modify their physical properties. To date, there are no enzymes that have completely degraded GA | + | Gum Arabic (GA) is a representative protein of the family of arabinogalactan proteins (AGPs) and is produced in acacia trees in response to stress conditions, such as drought or wounds. GA has a variety of applications within the industrial world, including the food, cosmetic, and pharmaceutical industries, acting specifically as an emulsion stabilizer, emulsifier, and thickener in pharmaceutical settings. However, the the detailed structure of GA has not determined because of the complex branching that occurs in the polysaccharide. Enzymes that can react with and eliminate glycosidic linkages of carbohydrates are useful for determining the structure and function of these carbohydrates, giving researchers the opportunity to modify their physical properties. To date, there are no enzymes that have completely degraded GA <ref name="Kondo" />. |
== Important amino acids== | == Important amino acids== | ||
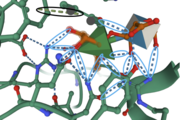
Amino Acids <scene name='89/892735/Amino_acids2/2'>His85, Asp106, Tyr150, Ser170, Tyr202, Arg220, Pro223, Ala225, and Arg331</scene> provide important interactions for binding in the active site. The amino acid residue <scene name='89/892735/His85interactions/1'>His85</scene> is located near the C-5 atom of Rha, suggesting it has a catalytic role as a proton acceptor within the active site. The <scene name='89/892735/His85_and_his105/1'>His105 side chain forms a hydrogen bond with His85</scene> side chain, providing stabilizing assistance. <ref name="Kondo">PMID:34303708</ref>. Amino Acid Residues <scene name='89/892735/Tyr150_and_his85/1'>Tyr150 and His85</scene> form a hydrogen bond with the O-1 atom of Rha, suggesting these residues play a catalytic role for the elimination reaction. <scene name='89/892735/O2o3hbonds/1'>Tyr26, Asn275, Gln276, and Arg331</scene> form hydrogen bonds with O-2 and O-3 atoms of Rha, providing further stabilization within the active site.[[Image:IntermolforcesBIOCHEM.png|thumb]] | Amino Acids <scene name='89/892735/Amino_acids2/2'>His85, Asp106, Tyr150, Ser170, Tyr202, Arg220, Pro223, Ala225, and Arg331</scene> provide important interactions for binding in the active site. The amino acid residue <scene name='89/892735/His85interactions/1'>His85</scene> is located near the C-5 atom of Rha, suggesting it has a catalytic role as a proton acceptor within the active site. The <scene name='89/892735/His85_and_his105/1'>His105 side chain forms a hydrogen bond with His85</scene> side chain, providing stabilizing assistance. <ref name="Kondo">PMID:34303708</ref>. Amino Acid Residues <scene name='89/892735/Tyr150_and_his85/1'>Tyr150 and His85</scene> form a hydrogen bond with the O-1 atom of Rha, suggesting these residues play a catalytic role for the elimination reaction. <scene name='89/892735/O2o3hbonds/1'>Tyr26, Asn275, Gln276, and Arg331</scene> form hydrogen bonds with O-2 and O-3 atoms of Rha, providing further stabilization within the active site.[[Image:IntermolforcesBIOCHEM.png|thumb]] | ||
Revision as of 18:37, 8 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
Structure and Function of FoRham1
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Kondo T, Kichijo M, Maruta A, Nakaya M, Takenaka S, Arakawa T, Fushinobu S, Sakamoto T. Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family. J Biol Chem. 2021 Jul 22:101001. doi: 10.1016/j.jbc.2021.101001. PMID:34303708 doi:http://dx.doi.org/10.1016/j.jbc.2021.101001