We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1694
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Important amino acids== | == Important amino acids== | ||
[[Image:DPIDxT.png | thumb]] | [[Image:DPIDxT.png | thumb]] | ||
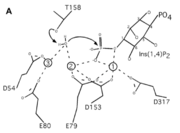
| - | INPP1D54A does not have a catalytic triad though it has six important <scene name='89/892737/Catalytic_amino_acids/1'>catalytic amino acids</scene> (D54, E80, E79, D153, D317, T158) that promote catalysis in the reaction along with the metal ions that helped the protein function. These amino acids hold the metal ions in place and metal ions hold on to the water and substrate. Thr158 is a very important catalytic amino acid because it is going to do a nucleophilic attack on the water molecule (shown in the picture on the right) which will result in a cascade of other reactions to happen (series of bonds breaking and being made) eventually leading to a phosphate leaving group. Hence, why the function of the enzyme is to remove a phosphate group | + | INPP1D54A does not have a catalytic triad though it has six important <scene name='89/892737/Catalytic_amino_acids/1'>catalytic amino acids</scene> (D54, E80, E79, D153, D317, T158) that promote catalysis in the reaction along with the metal ions that helped the protein function. These amino acids hold the metal ions in place and metal ions hold on to the water and substrate. Thr158 is a very important catalytic amino acid because it is going to do a nucleophilic attack on the water molecule (shown in the picture on the right) which will result in a cascade of other reactions to happen (series of bonds breaking and being made) eventually leading to a phosphate leaving group. Hence, why the function of the enzyme is to remove a phosphate group. |
| Line 31: | Line 31: | ||
In the structure INPP1D54A, there is a mutation in the amino acid aspartic acid (D)54 and causes it to change to alanine (A)54 (<scene name='89/892737/Alanine/1'>mutation in D54</scene>). This mutation does not impact the substrate affinity but does decrease the activity of INPP1. <ref name="dollins" /> | In the structure INPP1D54A, there is a mutation in the amino acid aspartic acid (D)54 and causes it to change to alanine (A)54 (<scene name='89/892737/Alanine/1'>mutation in D54</scene>). This mutation does not impact the substrate affinity but does decrease the activity of INPP1. <ref name="dollins" /> | ||
| - | + | [[Image:Motif_Copy.png | thumb]] | |
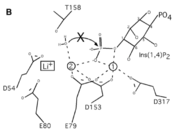
| - | A six amino acid <scene name='89/892737/Motif/1'>motif</scene>, DPIDxT anchors the metal-binding sites in the protein that are likely involved in catalysis while the metal binds to the substrate. <ref name="dollins" /> The sixth amino acid, x, is not as important as the other five, however, it can be any amino acid depending on the related crystallized structure to INPP1D54A or to a similar protein. | + | A six amino acid <scene name='89/892737/Motif/1'>motif</scene>, DPIDxT anchors the metal-binding sites in the protein that are likely involved in catalysis while the metal binds to the substrate. <ref name="dollins" /> The sixth amino acid, x, is not as important as the other five, however, it can be any amino acid depending on the related crystallized structure to INPP1D54A or to a similar protein. Also, lithium is an uncompetitive inhibitor for this protein and when it binds to a metal site (metal site 3 in the lower image B) it causes the protein not to function properly. |
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references /> | <references /> | ||
Revision as of 14:59, 9 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
Inositol polyphosphate 1-Phosphatase (INPP1) D54A
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ 3.0 3.1 3.2 Dollins DE, Xiong JP, Endo-Streeter S, Anderson DE, Bansal VS, Ponder JW, Ren Y, York JD. A Structural Basis for Lithium and Substrate Binding of an Inositide Phosphatase. J Biol Chem. 2020 Nov 10. pii: RA120.014057. doi: 10.1074/jbc.RA120.014057. PMID:33172890 doi:http://dx.doi.org/10.1074/jbc.RA120.014057