User:Mathias Bortoletto Dunker/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 61: | Line 61: | ||
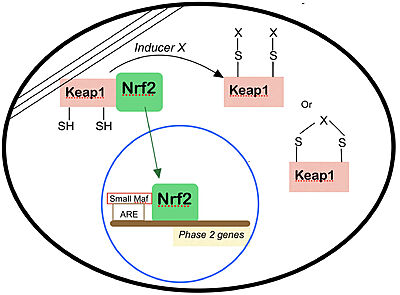
Thanks to the many binding sites of Nrf2, studies have shown that there are many alternative pathway from which Nrf2 degradation can occur, providing many mechanism for cellular control of Nrf2 activities <ref>Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034</ref>, subjected to Keap1 regulation or otherwise. The following diagram from Warren L. Wu and Thales Papagiannakopoulos <ref>https://www.annualreviews.org/doi/full/10.1146/annurev-cancerbio-030518-055627, CC BY 4.0, https://commons.wikimedia.org/w/index.php?curid=94063579</ref> summarizes all known Nrf2 related metabolic processes. | Thanks to the many binding sites of Nrf2, studies have shown that there are many alternative pathway from which Nrf2 degradation can occur, providing many mechanism for cellular control of Nrf2 activities <ref>Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034</ref>, subjected to Keap1 regulation or otherwise. The following diagram from Warren L. Wu and Thales Papagiannakopoulos <ref>https://www.annualreviews.org/doi/full/10.1146/annurev-cancerbio-030518-055627, CC BY 4.0, https://commons.wikimedia.org/w/index.php?curid=94063579</ref> summarizes all known Nrf2 related metabolic processes. | ||
| - | [[Image:Inputs_and_outputs_of_KEAP1_NRF2_pathway.jpg| | + | [[Image:Inputs_and_outputs_of_KEAP1_NRF2_pathway.jpg|600px|center|]] |
== Structural and Genetic Information of Nrf2 == | == Structural and Genetic Information of Nrf2 == | ||
Revision as of 05:05, 13 December 2021
Keap1-Nrf2 Complex
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Lo, S.-C., Li, X., Henzl, M.T., Beamer, L.J. and Hannink, M. (2006), Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. The EMBO Journal, 25: 3605-3617. https://doi.org/10.1038/sj.emboj.7601243
- ↑ Pitoniak, A., & Bohmann, D. (2015). Mechanisms and functions of Nrf2 signaling in Drosophila. Free radical biology & medicine, 88(Pt B), 302–313. https://doi.org/10.1016/j.freeradbiomed.2015.06.020
- ↑ Data available in https://www.ncbi.nlm.nih.gov/gene/9817
- ↑ Mathers J, Fraser JA, McMahon M, Saunders RD, Hayes JD, McLellan LI. Antioxidant and cytoprotective responses to redox stress. Biochem Soc Symp. 2004;(71):157-76. doi: 10.1042/bss0710157. PMID: 15777020.
- ↑ Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001 Jun 2;477(1-2):7-21. doi: 10.1016/s0027-5107(01)00091-4. PMID: 11376682.
- ↑ Ceconi C, Boraso A, Cargnoni A, Ferrari R. Oxidative stress in cardiovascular disease: myth or fact? Arch Biochem Biophys. 2003 Dec 15;420(2):217-21. doi: 10.1016/j.abb.2003.06.002. PMID: 14654060.
- ↑ Leung L, Kwong M, Hou S, Lee C, Chan JY. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J Biol Chem. 2003 Nov 28;278(48):48021-9. doi: 10.1074/jbc.M308439200. Epub 2003 Sep 10. PMID: 12968018.
- ↑ 8.0 8.1 Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11908-13. doi: 10.1073/pnas.172398899. Epub 2002 Aug 22. PMID: 12193649; PMCID: PMC129367.
- ↑ 9.0 9.1 9.2 Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004 Nov;10(11):549-57. doi: 10.1016/j.molmed.2004.09.003. PMID: 15519281.
- ↑ Sasaki, H. et al. (2002) Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 277, 44765–44771
- ↑ Chan, K. and Kan, Y.W. (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 96, 12731–12736
- ↑ Enomoto, A. et al. (2001) High sensitivity of Nrf2 knockout mice to Review TRENDS in Molecular Medicine Vol.10 No.11 November 2004 555 www.sciencedirect.com acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177
- ↑ Goldring, C.E. et al. (2004) Activation of hepatic Nrf2 in vivo by acetaminophen in CD-1 mice. Hepatology 39, 1267–1276
- ↑ Ramos-Gomez, M. et al. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 98, 3410–3415
- ↑ Cho, H-Y. et al. (2002) Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26, 175–182
- ↑ Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034
- ↑ https://www.annualreviews.org/doi/full/10.1146/annurev-cancerbio-030518-055627, CC BY 4.0, https://commons.wikimedia.org/w/index.php?curid=94063579
- ↑ Canning P, Sorrell FJ, Bullock AN. Structural basis of Keap1 interactions with Nrf2. Free Radic Biol Med. 2015;88(Pt B):101-107. doi:10.1016/j.freeradbiomed.2015.05.034