Sandbox Reserved 1647
From Proteopedia
| Line 1: | Line 1: | ||
<Structure load='5t1j' size='350' frame='true' align='right' caption='TBX21 on two DNA' scene='86/868180/Tbx21_on_dna/1' /> | <Structure load='5t1j' size='350' frame='true' align='right' caption='TBX21 on two DNA' scene='86/868180/Tbx21_on_dna/1' /> | ||
==Introduction== | ==Introduction== | ||
| - | TBX21 (for t-box transcription factor 21) or T-bet is a [https://en.wikipedia.org/wiki/Transcription_factor transcription factor], more precisely a [[T-box | + | TBX21 (for t-box transcription factor 21) or T-bet is a [https://en.wikipedia.org/wiki/Transcription_factor transcription factor], more precisely a [[T-box protein]]. The DNA binding domain of T-bet has a dimer structure composes of two identical chains (<scene name='86/868180/Chain_a/2'>A</scene> / <scene name='86/868180/Chain_b/2'>B</scene> ) with a total molecular weight of 76,37 kDA. This binding domain allows the protein to bind itself on the DNA promotor or regulator area. T-box protein is able to <scene name='86/868180/Link_to_2_dna/1'>link two DNA molecules with only one dimer</scene>. Moreover, it is able to link two areas of the same DNA molecules which are far from each other and thus create a chromatin loop. |
The main function of this transcription factor is to direct [https://en.wikipedia.org/wiki/T_helper_cell TH1 cells differentiation] from naive CD4+ cells. Nevertheless, the action field of T-bet is larger than classical T-box protein. | The main function of this transcription factor is to direct [https://en.wikipedia.org/wiki/T_helper_cell TH1 cells differentiation] from naive CD4+ cells. Nevertheless, the action field of T-bet is larger than classical T-box protein. | ||
Revision as of 14:13, 28 December 2021
|
Contents |
Introduction
TBX21 (for t-box transcription factor 21) or T-bet is a transcription factor, more precisely a T-box protein. The DNA binding domain of T-bet has a dimer structure composes of two identical chains ( / ) with a total molecular weight of 76,37 kDA. This binding domain allows the protein to bind itself on the DNA promotor or regulator area. T-box protein is able to . Moreover, it is able to link two areas of the same DNA molecules which are far from each other and thus create a chromatin loop. The main function of this transcription factor is to direct TH1 cells differentiation from naive CD4+ cells. Nevertheless, the action field of T-bet is larger than classical T-box protein.
Location
TBX21 protein is coded by the TBX21 gene on the chromosome 17 in humans. More precisely on the band 17q21.32.
Structure and structural interactions
Primary Structure
The DNA binding domain of T-bet from Mus musculus for a monomer is located between the residues and
Secondary structure and interactions
The secondary structure of the protein allows it to bind with the DNA: The T-box domain consists of several repeats of and is involved in both dimerization and DNA binding. Thanks to some post-translational modifications of the protein’s residues, the transcription factor TBX21 can bind with DNA between the residues and and some proteins. Firstly, the ubiquitination of the residue allows TBX21 to bind with the DNA sequence. Lys-313 was lately found as a key site required for T-bet to interact with the IFN-γ gene promoter and to assure phosphorylation at Thr-302 [1]. Secondly, the phosphorylation of some residues allows TBX21 to interact with several proteins. For example, phosphorylation of allows TBX21 to interact with NFAT, the one of with RUNX1, that of with NF-кB p65 and finally the one of Y525 allows the interaction with GATA-3. [2]
Biological roles of TBX21
Regulation of Th cells differentiation by TBX21
The transcription factor T-bet directs Th1 cell differentiation. The molecular mechanisms that underlie this lineage-specific gene regulation are not completely understood but several hypotheses have already been made on the action's mechanism of T-bet. [3] We know that T-bet initiates Th1 lineage development from naive Thp cells by activating Th1 genetics and repressing the opposing Th2 programs. Th1 cells stimulate cellular immune response while Th2 stimulates humoral immune response and induces antibody production. [4]
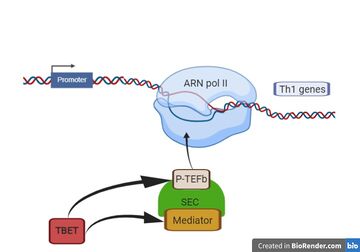
Here, we show that T-bet acts through enhancers to allow the recruitment of Mediator and P-TEFb in the formation of the super elongation complex (SEC). Th1 genes are occupied by RNA Polymerase II in Thp cells, while T-bet-mediated recruitment of P-TEFb and mediator. The recruitment of P-TEFb and mediator activates transcriptional elongation and giving place to increased differentiation of Thp into Th1.[5]
T-bet can also regulate Th1 cell differentiation by directly initiating gamma interferon (IFN-γ) transcription and by suppressing Th2-specific transcription factor GATA-3[6]. The T-bet induced expression of IFN-γ derives Th precursor cells to differentiate into Th1 effector cells. This stimulation of IFN-γ can take place thanks to the action of a nuclear tyrosine kinase, c-Abl. C-Abl induces phosphorylation of T-bet at tyrosine residues , , and . C-Abl phosphorylates the tyrosine residues within the T-box domain, which is the DNA-binding domain of T-bet. This phosphorylation leads to conformational changes of the T-box domain to facilitate the DNA-binding activity of T-bet and appears to play a crucial role in the IFN-γ promoter-binding activity of T-bet.
Recently, many studies have reported that T-bet also modulates other Th cell lineages, including Th17, Treg, and follicular Th (TFH) cells, in coordination with many transcription factors, such as the retinoic acid-related orphan receptor-𝛾t (ROR𝛾t) [7], runt-related transcription factor 3 (RUNX3), and B-cell lymphoma-6 (BCL6). These findings suggest that T-bet is a transcription factor that is critical for fine-tuning Th cell development.
TBX21 as an antiasthmatic regulator
Asthma remains one of the commonest chronic inflammatory diseases and has a major impact on the life of sufferers. It is associated with allergy mediated by IgE antibodies. T-bet was found associated with many immune-mediated diseases such as asthma.[8]
In asthmatic airways, Th2 cells are activated and release several cytokines [9] that regulate IgE production and inflammatory cell recruitment, such as eosinophils. Th2 cells and GATA-3 play an important role in allergic inflammation and asthma, and induce IgE production. The asthmatic patients present high levels of total IgE. On the contrary, the T-bet gene expression and Th1 pattern, along with the IFN- γ production, are usually associated with non-allergic asthmatics and healthy subjects.[8]
In T-bet structure, ubiquitination takes place at . It has an impact on the stability of the protein and leads to the degradation of the protein by the proteosome. Some research found the role of deubiquitinases involved in T-bet stability and function. As a deubiquitinase, USP10 belongs to the ubiquitin-specificprotease family of cysteine proteases. Cysteine 424 site on USP10 is crucial for its hydrolase activity. Results have shown that USP10 could interact with T-bet and stabilize it via interaction between Lysine 313 (K313) of T-bet and Cysteine 424 of USP10. Deubiquitination inhibits its degradation by the proteosome and enhance the secretion of IFN- γ.
Researchers believe that the USP10-dependent T-bet deubiquitination and stabilization can regulate antigen induced immune disorder especially in Th1 specific inflammation. Thus, appropriate decreasing USP10 level may contribute to the T-bet degradation and inflammation attenuation. [10]
References
- ↑ Mehta DS, Wurster AL, Weinmann AS, Grusby MJ. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc Natl Acad Sci U S A. 2005 Feb 8;102(6):2016-21. doi:, 10.1073/pnas.0409512102. Epub 2005 Jan 31. PMID:15684054 doi:http://dx.doi.org/10.1073/pnas.0409512102
- ↑ Jang EJ, Park HR, Hong JH, Hwang ES. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. J Immunol. 2013 Jun 1;190(11):5764-70. doi: 10.4049/jimmunol.1203403. Epub 2013, Apr 24. PMID:23616576 doi:http://dx.doi.org/10.4049/jimmunol.1203403
- ↑ Wang P, Wang Y, Xie L, Xiao M, Wu J, Xu L, Bai Q, Hao Y, Huang Q, Chen X, He R, Li B, Yang S, Chen Y, Wu Y, Ye L. The Transcription Factor T-Bet Is Required for Optimal Type I Follicular Helper T Cell Maintenance During Acute Viral Infection. Front Immunol. 2019 Mar 29;10:606. doi: 10.3389/fimmu.2019.00606. eCollection, 2019. PMID:30984183 doi:http://dx.doi.org/10.3389/fimmu.2019.00606
- ↑ Hertweck A, Evans CM, Eskandarpour M, Lau JC, Oleinika K, Jackson I, Kelly A, Ambrose J, Adamson P, Cousins DJ, Lavender P, Calder VL, Lord GM, Jenner RG. T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex. Cell Rep. 2016 Jun 21;15(12):2756-70. doi: 10.1016/j.celrep.2016.05.054. Epub, 2016 Jun 9. PMID:27292648 doi:http://dx.doi.org/10.1016/j.celrep.2016.05.054
- ↑ doi: https://dx.doi.org/10.1016/s0092-8674(00)80702-3
- ↑ Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011 Jan;12(1):96-104. doi: 10.1038/ni.1969. Epub 2010 Dec 12. PMID:21151104 doi:http://dx.doi.org/10.1038/ni.1969
- ↑ 8.0 8.1 doi: https://dx.doi.org/10.1016/s0960-9822(02)00830-8
- ↑ Oh S, Hwang ES. The role of protein modifications of T-bet in cytokine production and differentiation of T helper cells. J Immunol Res. 2014;2014:589672. doi: 10.1155/2014/589672. Epub 2014 May 13. PMID:24901011 doi:http://dx.doi.org/10.1155/2014/589672
- ↑ Pan L, Chen Z, Wang L, Chen C, Li D, Wan H, Li B, Shi G. Deubiquitination and stabilization of T-bet by USP10. Biochem Biophys Res Commun. 2014 Jul 4;449(3):289-94. doi:, 10.1016/j.bbrc.2014.05.037. Epub 2014 May 17. PMID:24845384 doi:http://dx.doi.org/10.1016/j.bbrc.2014.05.037
1 Mehta, D. S., Wurster, A. L., Weinmann, A. S., & Grusby, M. J. (2005). NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proceedings of the National Academy of Sciences of the United States of America, 102(6), 2016–2021. https://doi.org/10.1073/pnas.0409512102
2 Lysine 313 of T-box Is Crucial for Modulation of Protein Stability, DNA Binding, and Threonine Phosphorylation of T-bet Eun Jung Jang, Hye Ryeon Park, Jeong-Ho Hong, Eun Sook Hwang The Journal of Immunology June 1, 2013, 190 (11) 5764-5770; DOI: 10.4049/jimmunol.1203403
3 Oh, S., & Hwang, E. S. (2014). The role of protein modifications of T-bet in cytokine production and differentiation of T helper cells. Journal of immunology research, 2014, 589672. https://doi.org/10.1155/2014/589672
4 Pan, L., Chen, Z., Wang, L., Chen, C., Li, D., Wan, H., Li, B., & Shi, G. (2014). Deubiquitination and stabilization of T-bet by USP10. Biochemical and biophysical research communications, 449(3), 289–294. https://doi.org/10.1016/j.bbrc.2014.05.037
5 Wang, P., Wang, Y., Xie, L., Xiao, M., Wu, J., Xu, L., Bai, Q., Hao, Y., Huang, Q., Chen, X., He, R., Li, B., Yang, S., Chen, Y., Wu, Y., & Ye, L. (2019). The Transcription Factor T-Bet Is Required for Optimal Type I Follicular Helper T Cell Maintenance During Acute Viral Infection. Frontiers in immunology, 10, 606. https://doi.org/10.3389/fimmu.2019.00606
6 Hertweck, A., Evans, C. M., Eskandarpour, M., Lau, J. C., Oleinika, K., Jackson, I., Kelly, A., Ambrose, J., Adamson, P., Cousins, D. J., Lavender, P., Calder, V. L., Lord, G. M., & Jenner, R. G. (2016). T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex. Cell reports, 15(12), 2756–2770. https://doi.org/10.1016/j.celrep.2016.05.054
7 Koch, M. A., Tucker-Heard, G., Perdue, N. R., Killebrew, J. R., Urdahl, K. B., & Campbell, D. J. (2009). The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology, 10(6), 595–602. https://doi.org/10.1038/ni.1731
8 Lazarevic, V., Chen, X., Shim, J. H., Hwang, E. S., Jang, E., Bolm, A. N., Oukka, M., Kuchroo, V. K., & Glimcher, L. H. (2011). T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nature immunology, 12(1), 96–104. https://doi.org/10.1038/ni.1969
9 Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G., & Glimcher, L. H. (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell, 100(6), 655–669. https://doi.org/10.1016/s0092-8674(00)80702-3
10 Jenner, R. G., Townsend, M. J., Jackson, I., Sun, K., Bouwman, R. D., Young, R. A., Glimcher, L. H., & Lord, G. M. (2009). The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proceedings of the National Academy of Sciences of the United States of America, 106(42), 17876–17881. https://doi.org/10.1073/pnas.0909357106