FirstGlance/Visualizing Conservation
From Proteopedia
(→Initial View of Conservation) |
(→Initial View of Conservation) |
||
| Line 13: | Line 13: | ||
==Initial View of Conservation== | ==Initial View of Conservation== | ||
Initially, FirstGlance colors each amino acid in the protein chain processed by ConSurf according to its level of conservation. This gives you an overview, enabling you to see patches of highly conserved residues (functional sites). | Initially, FirstGlance colors each amino acid in the protein chain processed by ConSurf according to its level of conservation. This gives you an overview, enabling you to see patches of highly conserved residues (functional sites). | ||
| + | |||
| + | ===Multiple Chains=== | ||
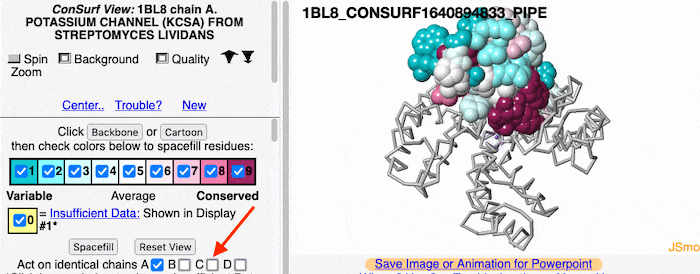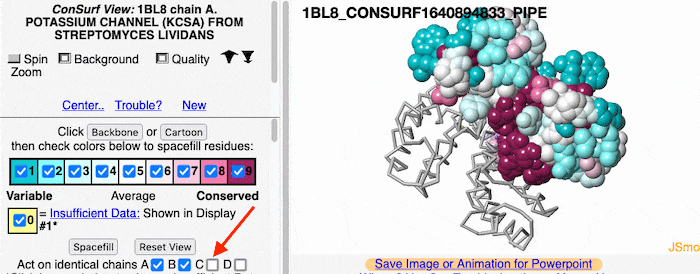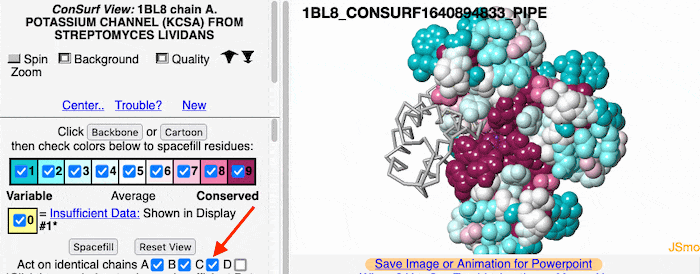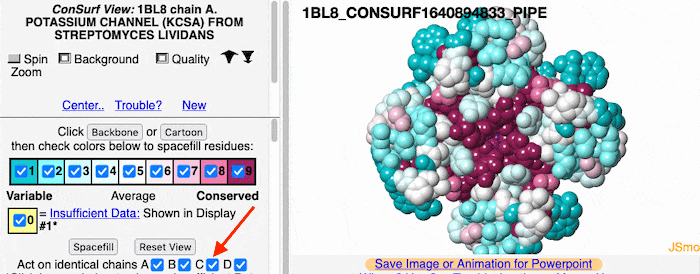
If the model has more than one copy of the chain processed by ConSurf, you can apply conservation colors to all of those sequence-identical chains. Here, for example, is a potassium channel homotetramer: | If the model has more than one copy of the chain processed by ConSurf, you can apply conservation colors to all of those sequence-identical chains. Here, for example, is a potassium channel homotetramer: | ||
Revision as of 18:09, 2 January 2022
This page is under construction. It is incomplete. Eric Martz 01:22, 1 January 2022 (UTC)
FirstGlance in Jmol is the easiest-to-use tool for visualizing, understanding, and sharing protein molecular structure. It offers guided exploration with extensive explanations and examples, yet provides ample power for advanced users. With a few mouse clicks, any molecular scene can be saved as a slide presentation-ready animation (GIF file) -- see examples below, and tinyurl.com/movingmolecules.
Among many conveniences offered by FirstGlance, it is easy to see the levels of evolutionary conservation determined by ConSurf for amino acids of special interest. Below is an overview with some illustrated examples. Elsewhere you will find instructions for running ConSurf on proteins of interest.
|
IMPORTANT (January, 2022): Use the unreleased beta-test version FirstGlance 3.8 Beta2 which has many improvements for ConSurf results beyond the publicly available version 3.7. Some of the features shown below are absent in version 3.7.
|
Contents |
Initial View of Conservation
Initially, FirstGlance colors each amino acid in the protein chain processed by ConSurf according to its level of conservation. This gives you an overview, enabling you to see patches of highly conserved residues (functional sites).
Multiple Chains
If the model has more than one copy of the chain processed by ConSurf, you can apply conservation colors to all of those sequence-identical chains. Here, for example, is a potassium channel homotetramer: