FirstGlance/Visualizing Conservation
From Proteopedia
(→Conservation of Salt Bridges) |
(→Conclusion) |
||
| Line 74: | Line 74: | ||
===Conclusion=== | ===Conclusion=== | ||
| - | + | Conservation of residues engaged in salt bridges varies over the entire range of conservation grades from 1 to 9. In some bridges, both residues are highly conserved (grades 8 or 9). In others, one partner is highly conserved, while the other is not. In some pairs, neither partner is conserved. | |
==Conservation of Cation-Pi Interactions== | ==Conservation of Cation-Pi Interactions== | ||
Revision as of 18:58, 3 January 2022
This page is under construction. It is incomplete. Eric Martz 01:22, 1 January 2022 (UTC)
FirstGlance in Jmol is the easiest-to-use tool for visualizing, understanding, and sharing protein molecular structure. It offers guided exploration with extensive explanations and examples, yet provides ample power for advanced users. With a few mouse clicks, any molecular scene can be saved as a slide presentation-ready animation (GIF file) -- see examples below, and tinyurl.com/movingmolecules.
Among many conveniences offered by FirstGlance, it is easy to see the levels of evolutionary conservation determined by ConSurf for amino acids of special interest. Below is an overview with some illustrated examples. Elsewhere you will find instructions for running ConSurf on proteins of interest.
|
IMPORTANT (January, 2022): Use the unreleased beta-test version FirstGlance 3.8 Beta2 which has many improvements for ConSurf results beyond the publicly available version 3.7. Some of the features shown below are absent in version 3.7. If you wish to try out the examples used below, click
At your finished ConSurf Job Status page, under the heading PDB Files, right click on PDB File with ConSurf Results in its Header, for FirstGlance in Jmol and select Copy Link Address. Then, at FirstGlance 3.8 Beta2, click enter a molecule's URL, paste the address into the slot, and click Submit. (Uploading the downloaded ConSurf PDB file won't work, because the upload mechanism always goes to version 3.7.) |
Initial View of Conservation
Initially, FirstGlance colors each amino acid in the protein chain processed by ConSurf according to its level of conservation. This gives you an overview, enabling you to see patches of highly conserved residues (functional sites).
Multiple Chains
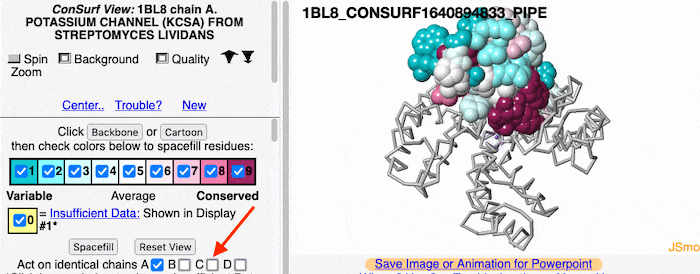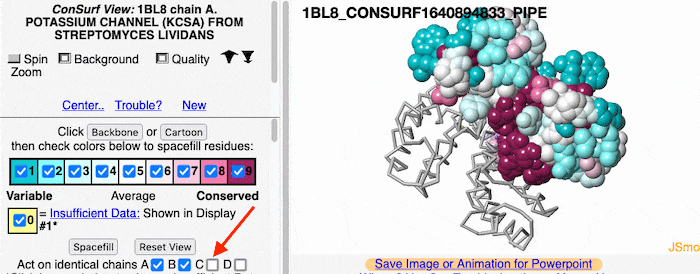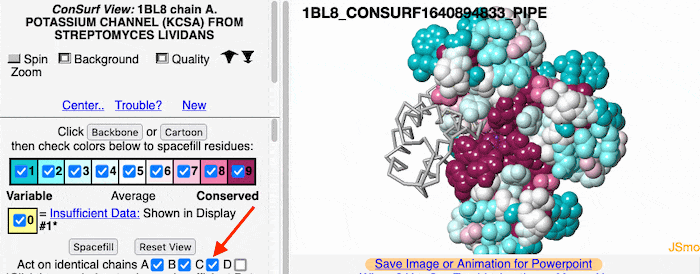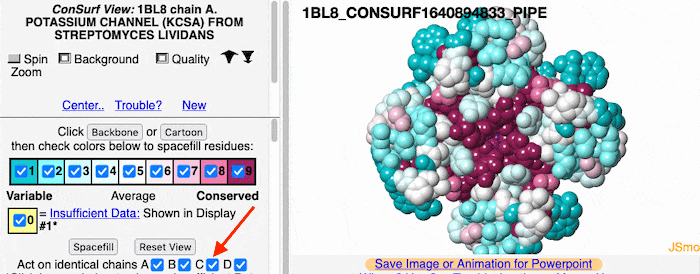
If the model has more than one copy of the chain processed by ConSurf, you can apply conservation colors to all of those sequence-identical chains. Here, for example, is a potassium channel homotetramer:
Optimization
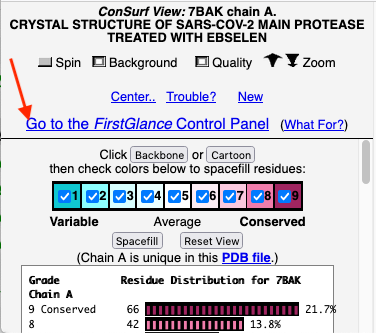
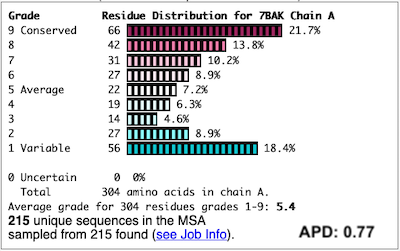
Before going further, it is good to decide whether the results are satisfactory, or need optimization. Lets consider 7bak, a drug-target protease of SARS-CoV-2 (see Coronavirus Proteases). A ConSurf run with all default settings left 10% of the residues with insufficient data[1] (yellow color), and the diversity in the multiple sequence alignment (MSA) was too low (average pairwise distance 0.34), leaving conservation grades 2 and 3 almost unoccupied. These inadequacies are highlighted by a distribution report displayed by FirstGlance.
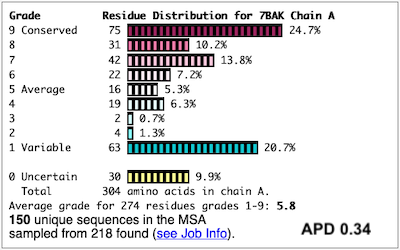
To improve the results, a new ConSurf job for 7bak was submitted, increasing the HMMER search iterations from 1 to 2, and increasing the maximum number of sequences in the MSA from 150 to 300. The results were satisfactory, with a more reasonable APD of 0.77.
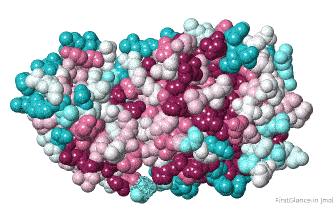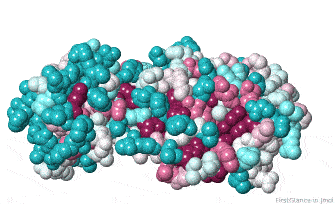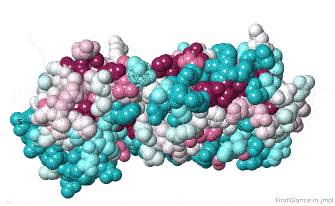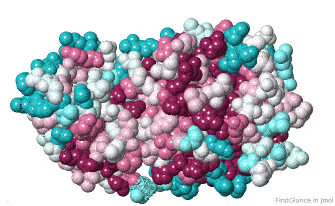
Initial View: Conserved Face
The initial view utilizes the "ConSurf Control Panel" (labeled ConSurf View). One side of the molecule is more conserved than the opposite side. The more conserved face includes the catalytic site, as we shall see.
Change Control Panels
To further explore the pattern of conservation, we need to change from the ConSurf View to the FirstGlance Control Panel. Click the link indicated by the red arrow in the above snapshot.
The FirstGlance Control Panel has 5 tabs. The Molecule Information Tab and the Views tab provide a wealth of information, but not related to conservation. For the purposes of this page, we'll skip over them. Notice that you can move freely back and forth between the ConSurf Controls and the FirstGlance Controls using the link ConSurf Controls in the middle of the left side of FirstGlance.
Conservation of Salt Bridges
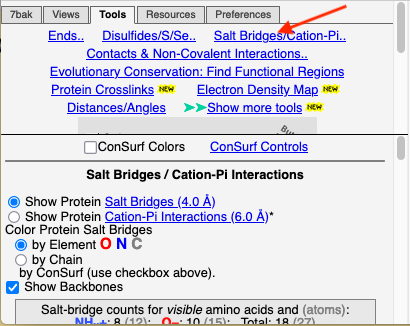
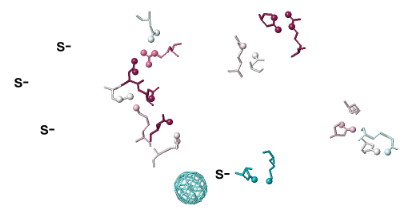
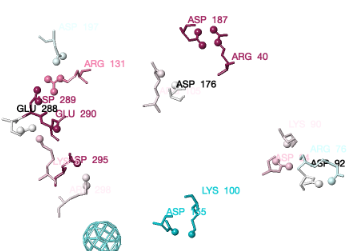
Click the Tools tab, and there click Salt Bridges/Cation-Pi.. (red arrow below). This shows all the salt bridges in the model where Negative (Asp, Glu) charges are within 4.0 Å of Positive (Arg, Lys) charges.
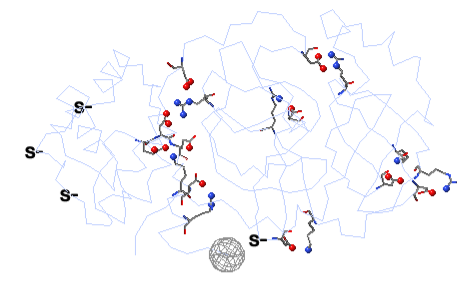
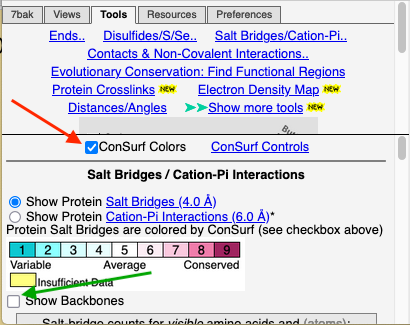
To see how well the salt-bridged residues are conserved, click the checkbox ConSurf Colors (red arrow below). Declutter the view by unchecking Show Backbones (green arrow below).
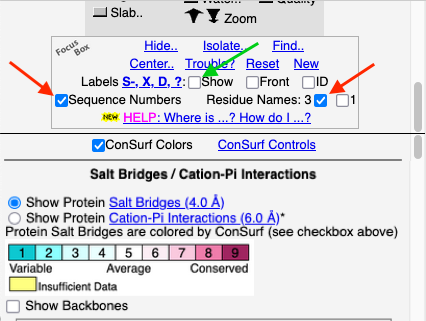
You can identify residues by touching them (a tooltip appears), or clicking on them. But it may be more convenient to label all salt bridge residues. Scroll down to the Focus Box in the upper left panel. Here you can check Sequence Numbers and Residue Names (red arrows below). If you wish, you can hide the S- labels that designate incomplete sidechains by unchecking the checkbox Labels, Show (green arrow below).
Conclusion
Conservation of residues engaged in salt bridges varies over the entire range of conservation grades from 1 to 9. In some bridges, both residues are highly conserved (grades 8 or 9). In others, one partner is highly conserved, while the other is not. In some pairs, neither partner is conserved.
Conservation of Cation-Pi Interactions
This can be done the same way as Salt Bridges, just clicking the radio button to switch from Salt Bridges to Cation-Pi interactions (see snapshots above).
Conservation of Residues Binding Ligand/Substrate
For this example, we'll use acetylcholinesterase complexed to the inhibitor VX, crystal structure 6cqz. From the ConSurf View, go to the FirstGlance Control Panel. FirstGlance provides an easy and powerful tool to explore residues contacting any specified portion of the model. In the Tools tab, click Contacts & Non-Covalent Interactions...
Conservation of Protein Crosslinks
INCLUDING SALT BRIDGES (ACHE)
Conservation of Specified Residues
See Also
References
- ↑ When the confidence interval for a conservation value of an amino acid is too large, ConSurf reports "insufficient data" (uncertainty: yellow color). See Position Specific Quality and Confidence Interval in the ConSurf documentation.