We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1647
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
===Primary Structure=== | ===Primary Structure=== | ||
| - | The DNA binding domain of T-bet from ''Mus musculus'' for a monomer is located between the residues <scene name='86/868180/Lys135/1'>LYS 135</scene> and <scene name='86/868180/Asn_326/1'>ASN 326</scene> [[Image:T-Bet primary structure.PNG|thumb |left|upright=3 | | + | The DNA binding domain of T-bet from ''Mus musculus'' for a monomer is located between the residues <scene name='86/868180/Lys135/1'>LYS 135</scene> and <scene name='86/868180/Asn_326/1'>ASN 326</scene> [[Image:T-Bet primary structure.PNG|thumb |left|upright=3 |Figure of primary structure of T-bet protein from ''Mus musculus'']] |
Revision as of 12:59, 4 January 2022
| |||||||||||
References
- ↑ TBX21-GeneCards : TBX21 - T-box Transcription factor 21 :https://www.genecards.org/cgi-bin/carddisp.pl?gene=TBX21
- ↑ Mehta DS, Wurster AL, Weinmann AS, Grusby MJ. NFATc2 and T-bet contribute to T-helper-cell-subset-specific regulation of IL-21 expression. Proc Natl Acad Sci U S A. 2005 Feb 8;102(6):2016-21. doi:, 10.1073/pnas.0409512102. Epub 2005 Jan 31. PMID:15684054 doi:http://dx.doi.org/10.1073/pnas.0409512102
- ↑ Jang EJ, Park HR, Hong JH, Hwang ES. Lysine 313 of T-box is crucial for modulation of protein stability, DNA binding, and threonine phosphorylation of T-bet. J Immunol. 2013 Jun 1;190(11):5764-70. doi: 10.4049/jimmunol.1203403. Epub 2013, Apr 24. PMID:23616576 doi:http://dx.doi.org/10.4049/jimmunol.1203403
- ↑ Wang P, Wang Y, Xie L, Xiao M, Wu J, Xu L, Bai Q, Hao Y, Huang Q, Chen X, He R, Li B, Yang S, Chen Y, Wu Y, Ye L. The Transcription Factor T-Bet Is Required for Optimal Type I Follicular Helper T Cell Maintenance During Acute Viral Infection. Front Immunol. 2019 Mar 29;10:606. doi: 10.3389/fimmu.2019.00606. eCollection, 2019. PMID:30984183 doi:http://dx.doi.org/10.3389/fimmu.2019.00606
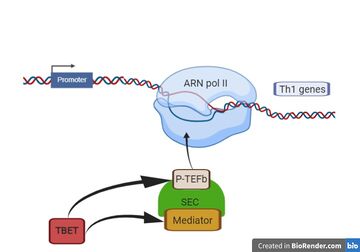
- ↑ Hertweck A, Evans CM, Eskandarpour M, Lau JC, Oleinika K, Jackson I, Kelly A, Ambrose J, Adamson P, Cousins DJ, Lavender P, Calder VL, Lord GM, Jenner RG. T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex. Cell Rep. 2016 Jun 21;15(12):2756-70. doi: 10.1016/j.celrep.2016.05.054. Epub, 2016 Jun 9. PMID:27292648 doi:http://dx.doi.org/10.1016/j.celrep.2016.05.054
- ↑ Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G., & Glimcher, L. H. (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell, 100(6), 655–669. https://doi.org/10.1016/s0092-8674(00)80702-3
- ↑ Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011 Jan;12(1):96-104. doi: 10.1038/ni.1969. Epub 2010 Dec 12. PMID:21151104 doi:http://dx.doi.org/10.1038/ni.1969
- ↑ 9.0 9.1 Douglas.S.Robinson and Clare M Lloyd. Asthma: T-bet - A master controller ? Volume 12, Issue 9, PR322-R324, April 30, (2002) https://doi.org/10.1016/S0960-9822(02)00830-8
- ↑ Oh S, Hwang ES. The role of protein modifications of T-bet in cytokine production and differentiation of T helper cells. J Immunol Res. 2014;2014:589672. doi: 10.1155/2014/589672. Epub 2014 May 13. PMID:24901011 doi:http://dx.doi.org/10.1155/2014/589672
- ↑ Pan L, Chen Z, Wang L, Chen C, Li D, Wan H, Li B, Shi G. Deubiquitination and stabilization of T-bet by USP10. Biochem Biophys Res Commun. 2014 Jul 4;449(3):289-94. doi:, 10.1016/j.bbrc.2014.05.037. Epub 2014 May 17. PMID:24845384 doi:http://dx.doi.org/10.1016/j.bbrc.2014.05.037
12. Koch, M. A., Tucker-Heard, G., Perdue, N. R., Killebrew, J. R., Urdahl, K. B., & Campbell, D. J. (2009). The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nature immunology, 10(6), 595–602. https://doi.org/10.1038/ni.1731