We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1659
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
<StructureSection load='2xl4' size='340' side='right' caption='Caption for this structure ' scene=''> | <StructureSection load='2xl4' size='340' side='right' caption='Caption for this structure ' scene=''> | ||
| Line 9: | Line 8: | ||
| - | == | + | == Function== |
| - | Once the | + | Once the bacteria reaches the host's cytoplasm, the expression of LntA is activated, the protein is excreted and addressed to the nucleus thanks to a peptide signal. Then, LntA interacts with the transcription factor [[BAHD1]] [https://pubmed.ncbi.nlm.nih.gov/19666599/]. In absence of infection, BAHD1 represses the expression of ISG by promoting the local formation of heterochromatin while the interaction of LntA with BAHD1 has the effect of removing the chromatin repressor from the host’s DNA. Therefore, L. monocytogenes virulence factor induces a strong interferon response which enhances its pathogenicity. |
The mechanisms by which Listeria benefits from the synthesis of interferons are not fully understood. One hypothesis could be that Listeria monocytogenes takes advantage of the arrest of cellular-cycle induced by interferons. <ref>ROHDE JOHN R. Listeria unwinds host’s DNA. SCIENCE, 2011 : 1271-1272</ref> Indeed, this mechanism could be similar to those used by other pathogens such as Salmonella <ref>Winter SE, Thiennimitr P et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010</ref> or Yersinia <ref>Dewoody, R., Merritt, P.M., Houppert, A.S. and Marketon, M.M. (2011), YopK regulates the Yersinia pestis type III secretion system from within host cells. Molecular Microbiology, 79: 1445-1461. https://doi.org/10.1111/j.1365-2958.2011.07534.x</ref> which are able to promote an inflammatory response in gut epithelium in order to facilitate their dissemination and colonization. | The mechanisms by which Listeria benefits from the synthesis of interferons are not fully understood. One hypothesis could be that Listeria monocytogenes takes advantage of the arrest of cellular-cycle induced by interferons. <ref>ROHDE JOHN R. Listeria unwinds host’s DNA. SCIENCE, 2011 : 1271-1272</ref> Indeed, this mechanism could be similar to those used by other pathogens such as Salmonella <ref>Winter SE, Thiennimitr P et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010</ref> or Yersinia <ref>Dewoody, R., Merritt, P.M., Houppert, A.S. and Marketon, M.M. (2011), YopK regulates the Yersinia pestis type III secretion system from within host cells. Molecular Microbiology, 79: 1445-1461. https://doi.org/10.1111/j.1365-2958.2011.07534.x</ref> which are able to promote an inflammatory response in gut epithelium in order to facilitate their dissemination and colonization. | ||
In addition, Lebreton et al showed that when listeria grows outside the cell, the transcription rate of LntA is almost null and that a constitutive expression of LntA has an antibacterial effect. Thus, the efficiency of LntA requires a precise temporal and quantitative regulation. | In addition, Lebreton et al showed that when listeria grows outside the cell, the transcription rate of LntA is almost null and that a constitutive expression of LntA has an antibacterial effect. Thus, the efficiency of LntA requires a precise temporal and quantitative regulation. | ||
| Line 17: | Line 16: | ||
| - | == | + | == Structure== |
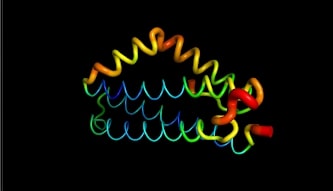
LntA is a small basic protein of 9.7 kDa. This protein is highly conserved in L. monocytogenes and is absent in almost all non-pathogenic Listeria strains . This characteristic suggests that lntA plays a key role in Listeria’s virulence. The acidic part of LntA is composed of aspartic acid (<scene name='86/868192/Acidic/1'>17,8%</scene>) and the basic part is composed of lysine and arginine (<scene name='86/868192/Basic/1'>18,6%</scene>). LntA is composed of 5 alpha-helix, three of them are long antiparallel helix and can be seen as the core of the protein. The two remaining helix stick out the core. The 3 first helix are named <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene>. The 2 others are <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene>. These residues located in <scene name='86/868192/These_two_helix/1'>these two helixes</scene> display high RMSD values meaning that this region is likely to oscillate. In fact, by studying this protein on pymol we can see it by the thickness and redness of these helixes which means that they have a high RMSD value : | LntA is a small basic protein of 9.7 kDa. This protein is highly conserved in L. monocytogenes and is absent in almost all non-pathogenic Listeria strains . This characteristic suggests that lntA plays a key role in Listeria’s virulence. The acidic part of LntA is composed of aspartic acid (<scene name='86/868192/Acidic/1'>17,8%</scene>) and the basic part is composed of lysine and arginine (<scene name='86/868192/Basic/1'>18,6%</scene>). LntA is composed of 5 alpha-helix, three of them are long antiparallel helix and can be seen as the core of the protein. The two remaining helix stick out the core. The 3 first helix are named <scene name='86/868192/Helix_h1/1'>H1</scene>, <scene name='86/868192/Helix_h2/1'>H2</scene> and <scene name='86/868192/Helix_h3/1'>H3</scene>. The 2 others are <scene name='86/868192/Helix_h4ter/1'>H4</scene> and <scene name='86/868192/Helix_h5/4'>H5</scene>. These residues located in <scene name='86/868192/These_two_helix/1'>these two helixes</scene> display high RMSD values meaning that this region is likely to oscillate. In fact, by studying this protein on pymol we can see it by the thickness and redness of these helixes which means that they have a high RMSD value : | ||
| Line 27: | Line 26: | ||
Nevertheless, the structure of the complex LntA-BAHD1 is not entirely resolved yet. <ref>Alice Lebreton. Régulations post-transcriptionnelles de l’expression génique de la cellule hôte en réponse à l’infection bactérienne. Sciences du Vivant, 2015</ref> | Nevertheless, the structure of the complex LntA-BAHD1 is not entirely resolved yet. <ref>Alice Lebreton. Régulations post-transcriptionnelles de l’expression génique de la cellule hôte en réponse à l’infection bactérienne. Sciences du Vivant, 2015</ref> | ||
| - | == | + | == Conclusion== |
The discovery of the LntA virulence factor shows that pathogenic bacteria can implement complex infectious strategies, requiring very precise temporal and quantitative regulation of the virulence factor delivery. Studies on virulence factors that target the nucleus can lead to the discovery of new mechanisms of gene expression regulation. And more importantly, the understanding these dialogs might allow new drug design and possibly a better support of the patients. Furthermore, the awareness of such mechanisms is very recent and raises many questions. For instance, it is still unclear whether the impact of lntA on gene expression is merely temporary or whether it leaves epigenetic marks over the long term. | The discovery of the LntA virulence factor shows that pathogenic bacteria can implement complex infectious strategies, requiring very precise temporal and quantitative regulation of the virulence factor delivery. Studies on virulence factors that target the nucleus can lead to the discovery of new mechanisms of gene expression regulation. And more importantly, the understanding these dialogs might allow new drug design and possibly a better support of the patients. Furthermore, the awareness of such mechanisms is very recent and raises many questions. For instance, it is still unclear whether the impact of lntA on gene expression is merely temporary or whether it leaves epigenetic marks over the long term. | ||
Revision as of 16:29, 19 January 2022
| |||||||||||
References
- ↑ ROHDE JOHN R. Listeria unwinds host’s DNA. SCIENCE, 2011 : 1271-1272
- ↑ Winter SE, Thiennimitr P et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010
- ↑ Dewoody, R., Merritt, P.M., Houppert, A.S. and Marketon, M.M. (2011), YopK regulates the Yersinia pestis type III secretion system from within host cells. Molecular Microbiology, 79: 1445-1461. https://doi.org/10.1111/j.1365-2958.2011.07534.x
- ↑ Lebreton A, Job V, Ragon M, Le Monnier A, Dessen A, Cossart P, Bierne H. 2014. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA
- ↑ Alice Lebreton. Régulations post-transcriptionnelles de l’expression génique de la cellule hôte en réponse à l’infection bactérienne. Sciences du Vivant, 2015