User:Ashley R. Wilkinson/Sandbox 1
From Proteopedia
| Line 6: | Line 6: | ||
==Overview of Structure== | ==Overview of Structure== | ||
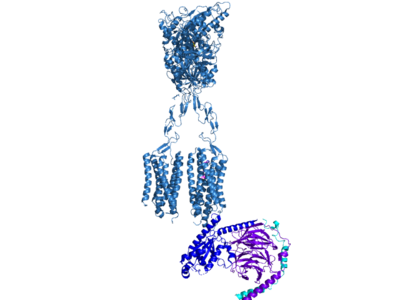
| - | Cryo-EM studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2 (Lin). The overall structure of the mGlu2 is composed of 3 main parts: a ligand binding Venus FlyTrap Domain(VFT), followed by a Cysteine Rich Domain linker to the Transmembrane Domain that contains 7 alpha helices (7TM) on both the <scene name='90/905587/Alphaandbetachain/ | + | Cryo-EM studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2 (Lin). The overall structure of the mGlu2 is composed of 3 main parts: a ligand binding Venus FlyTrap Domain(VFT), followed by a Cysteine Rich Domain linker to the Transmembrane Domain that contains 7 alpha helices (7TM) on both the <scene name='90/905587/Alphaandbetachain/2'>alpha and beta chain</scene>s that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. MGlu2 is a homodimer which is imperative to the receptor’s ability to relay signals induced by glutamate from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2. |
<StructureSection load='7mts' size='350' frame='true' align='right' caption='Metabotropic Glutamate Receptor'/> | <StructureSection load='7mts' size='350' frame='true' align='right' caption='Metabotropic Glutamate Receptor'/> | ||
Revision as of 19:25, 27 March 2022
Contents |
Metabotropic Glutamate Receptor mGluR-2
Introduction
Overview of Structure
Cryo-EM studies of mGlu2 have yielded adequate structures that have acted as maps to aid in producing a better structural understanding of the inactive and active states of mGlu2 (Lin). The overall structure of the mGlu2 is composed of 3 main parts: a ligand binding Venus FlyTrap Domain(VFT), followed by a Cysteine Rich Domain linker to the Transmembrane Domain that contains 7 alpha helices (7TM) on both the s that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. MGlu2 is a homodimer which is imperative to the receptor’s ability to relay signals induced by glutamate from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2.
|
This is a sentence about the structure of my protein. This is a sentence about the structure of my protein. This is a sentence about the structure of my protein. This is a sentence about the structure of my protein. This is a sentence about the structure of my protein. This is a sentence about the structure of my protein.
Function
Disease
Relevance
Structural highlights
This is my new image.This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
<7mts/>