We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1710
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
=== Open Conformation === | === Open Conformation === | ||
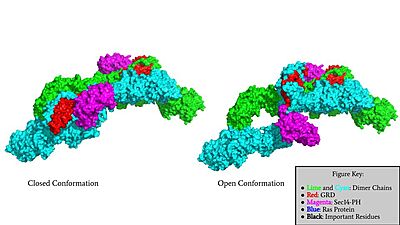
| - | The other conformation that characterizes Neurofibromin is the <scene name='90/904315/Open_conformation/2'>Open Conformation</scene>. In the open conformation of Neurofibromin, the protein is considered active and is participating in its function of Ras regulation. This occurs because the transition metal-binding site with zinc no longer is able to form due to an increase in distance between the C1032, H1558 and H1576 residues that form the <scene name='90/904315/Open_conformation_triade/3'>Open Triade</scene>. One protomer in Neurofibromin has its GRD and Sec14-PH domains oriented in way that is almost reversed in position compared to the closed conformation. Due to this rotation, C1032 is now located too far away, approximately 30 Angstroms, from H1558 and H1576 which results in the loss of the metal-binding site. The lack of the transition metal-binding site allows the GRD to orient itself in such a way that it can associate with <scene name='90/904315/Ras_open_conformation/1'>Ras in the Open Conformation</scene>. The reason that Neurofibromin is only able to associate with Ras in the open conformation is due to one critical residue, the <scene name='90/904315/Open_conformation_arginine_fin/1'>Arginine Finger</scene> located at position 1276 in Neurofibromin. When Neurofibromin is in the open conformation, R1276 is able to bind to Ras because there is no steric hindrance from the Neurofibromin core. | + | The other conformation that characterizes Neurofibromin is the <scene name='90/904315/Open_conformation/2'>Open Conformation</scene>. In the open conformation of Neurofibromin, the protein is considered active and is participating in its function of Ras regulation. This occurs because the transition metal-binding site with zinc no longer is able to form due to an increase in distance between the C1032, H1558 and H1576 residues that form the <scene name='90/904315/Open_conformation_triade/3'>Open Triade</scene>. One protomer in Neurofibromin has its GRD and Sec14-PH domains oriented in way that is almost reversed in position compared to the closed conformation. Due to this rotation, C1032 is now located too far away, approximately 30 Angstroms, from H1558 and H1576 which results in the loss of the metal-binding site. The lack of the transition metal-binding site allows the GRD to orient itself in such a way that it can associate with <scene name='90/904315/Ras_open_conformation/1'>Ras in the Open Conformation</scene>. The reason that Neurofibromin is only able to associate with Ras in the open conformation is due to one critical residue, the <scene name='90/904315/Open_conformation_arginine_fin/1'>Arginine Finger</scene> located at position 1276 in Neurofibromin. When Neurofibromin is in the open conformation, R1276 is able to <scene name='90/904315/Ras_open_conformation_with_arg/1'>bind to Ras</scene> because there is no steric hindrance from the Neurofibromin core. |
| - | *<scene name='90/904315/Ras_open_conformation_with_arg/1'>Ras with Open Conformation and Arginine Finger</scene> | ||
== Function == | == Function == | ||
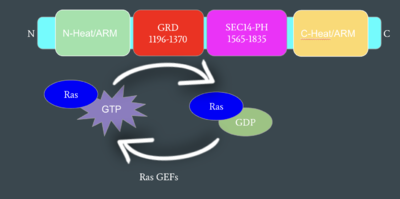
Neurofibromin functions as a tumor suppressor protein.<ref name="Trovó-Marqui">PMID:16813595</ref> Its job is to prevent cell growth by turning off another protein known as [https://en.wikipedia.org/wiki/Ras_GTPase Ras] which in its active state, stimulates cell growth and division. Ras is a GTPase membrane protein that can only interact with Neurofibromin, a cytoplasmic protein, in the open conformation. As Neurofibromin is a cytoplasmic protein, it is brought to the membrane to associate with Ras via another protein known as SPRED1. Neurofibromin can interact with SPRED1 in both the open and closed conformations however, it can only associate with Ras when it is in its open conformation. The interaction between Neurofibromin and Ras occurs via an arginine finger (R1276) present in the GRD of Neurofibromin which is critical for Ras binding. R1276 is only accessible for binding when the GRD and Sec14-PH domains of Neurofibromin are rotated into the open conformation and there is no steric hindrance from the surrounding dimer chains. When R1276 is able to associate with Ras, Neurofibromin downregulates the Ras signaling pathway by hydrolyzing the GTP associated with Ras to GDP, effectively making it inactive and inhibiting cell growth and division.[[Image:mechanismofRas.png|400 px|lef|thumb|Mechanism of Ras Regulation by Neurofibromin]] | Neurofibromin functions as a tumor suppressor protein.<ref name="Trovó-Marqui">PMID:16813595</ref> Its job is to prevent cell growth by turning off another protein known as [https://en.wikipedia.org/wiki/Ras_GTPase Ras] which in its active state, stimulates cell growth and division. Ras is a GTPase membrane protein that can only interact with Neurofibromin, a cytoplasmic protein, in the open conformation. As Neurofibromin is a cytoplasmic protein, it is brought to the membrane to associate with Ras via another protein known as SPRED1. Neurofibromin can interact with SPRED1 in both the open and closed conformations however, it can only associate with Ras when it is in its open conformation. The interaction between Neurofibromin and Ras occurs via an arginine finger (R1276) present in the GRD of Neurofibromin which is critical for Ras binding. R1276 is only accessible for binding when the GRD and Sec14-PH domains of Neurofibromin are rotated into the open conformation and there is no steric hindrance from the surrounding dimer chains. When R1276 is able to associate with Ras, Neurofibromin downregulates the Ras signaling pathway by hydrolyzing the GTP associated with Ras to GDP, effectively making it inactive and inhibiting cell growth and division.[[Image:mechanismofRas.png|400 px|lef|thumb|Mechanism of Ras Regulation by Neurofibromin]] | ||
Revision as of 19:38, 28 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Neurofibromin - The Tumor Suppressor Gene
| |||||||||||
References
- ↑ Naschberger A, Baradaran R, Rupp B, Carroni M. The structure of neurofibromin isoform 2 reveals different functional states. Nature. 2021 Nov;599(7884):315-319. doi: 10.1038/s41586-021-04024-x. Epub 2021, Oct 27. PMID:34707296 doi:http://dx.doi.org/10.1038/s41586-021-04024-x
- ↑ Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2