Sandbox Reserved 1700
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
=== Novel Characteristics === | === Novel Characteristics === | ||
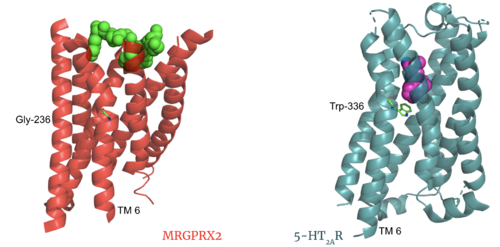
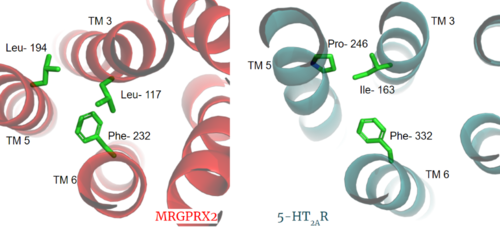
| - | The most important findings about the MRGPRX2 receptor are the differences between it and all other previously discovered GPCR's found in the A family. Many conserved structural motifs, characteristic of the A family receptors, are absent on MRGPRX2 | + | The most important findings about the MRGPRX2 receptor are the differences between it and all other previously discovered GPCR's found in the A family. Many conserved structural motifs, characteristic of the A family receptors, are absent on MRGPRX2. These structural motif differences contribute to a membrane surface ligand binding rather than a ligand binding deep within the helices. To demonstrate this difference in depth binding, MRGPRX2 is compared to [https://proteopedia.org/wiki/index.php/Serotonin_receptor 5-HT2AR], another class A GPCR with more conserved structural motifs. |
| + | ---- | ||
| + | ---- | ||
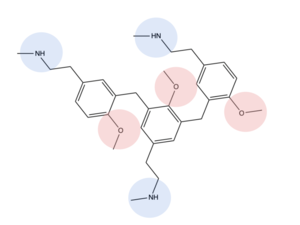
| + | [[Image:Screen Shot 2022-03-27 at 5.06.17 PM.png|500px|center|thumb|Comparison of ligand Cortistatin-14 binding in MRGPRX2 (left) and binding in 5HT2AR (right)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ---- | ||
| + | ---- | ||
<jmol> | <jmol> | ||
| Line 37: | Line 47: | ||
==== Sodium Site ==== | ==== Sodium Site ==== | ||
| - | + | The MRGPRX2 <scene name='90/904306/Sodium_site_2/1'>sodium binding site</scene> consists of ASP-75 and GLY-116 compared to the [https://proteopedia.org/wiki/index.php/Neurotensin_receptor#sodium%20binding%20pocket previously conserved] residues in this binding pocket. Other class A GPCRs demonstrate a larger binding pocket with a higher negative character allowing for a suitable environment for sodium ions to bind. In MRGPRX2, this pocket lacks the same amount of <scene name='90/904306/Sodium_site_charge/2'>negative character</scene> with the shift to a glycine residue rather than typical negative residues. The helices for the MRGPRX2 in the binding pocket are also more collapsed making this pocket less accessible for sodium ions. | |
==== PIF/LLF Motif ==== | ==== PIF/LLF Motif ==== | ||
| Line 52: | Line 62: | ||
==== DRY/ ERC Motif ==== | ==== DRY/ ERC Motif ==== | ||
| + | MRGPRX2 has an <scene name='90/904306/Erc_motif_3/1'>ERC Motif</scene> rather than the typically [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY Motif]. The amino acid residue shift from TYR-174 to CYS-128 has spatial arrangement implications where the helices are more compact in MRGPRX2 without the TYR to physically push the TMP helices apart. | ||
| + | [[Image:Screen Shot 2022-03-15 at 10.23.20 AM.png|200px|left|thumb|ERC Motif]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Revision as of 01:31, 29 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
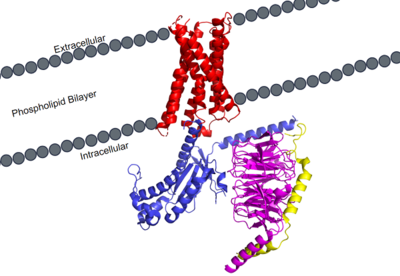
MRGPRX2 Human Itch G-Protein Coupled Receptor (GPCR)
| |||||||||||
References
- ↑ Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov. 2017 Dec;16(12):829-842. doi: 10.1038/nrd.2017.178. Epub, 2017 Oct 27. PMID:29075003 doi:http://dx.doi.org/10.1038/nrd.2017.178
- ↑ Basith S, Cui M, Macalino SJY, Park J, Clavio NAB, Kang S, Choi S. Exploring G Protein-Coupled Receptors (GPCRs) Ligand Space via Cheminformatics Approaches: Impact on Rational Drug Design. Front Pharmacol. 2018 Mar 9;9:128. doi: 10.3389/fphar.2018.00128. eCollection, 2018. PMID:29593527 doi:http://dx.doi.org/10.3389/fphar.2018.00128
- ↑ 3.0 3.1 3.2 3.3 3.4 Cao C, Kang HJ, Singh I, Chen H, Zhang C, Ye W, Hayes BW, Liu J, Gumpper RH, Bender BJ, Slocum ST, Krumm BE, Lansu K, McCorvy JD, Kroeze WK, English JG, DiBerto JF, Olsen RHJ, Huang XP, Zhang S, Liu Y, Kim K, Karpiak J, Jan LY, Abraham SN, Jin J, Shoichet BK, Fay JF, Roth BL. Structure, function and pharmacology of human itch GPCRs. Nature. 2021 Dec;600(7887):170-175. doi: 10.1038/s41586-021-04126-6. Epub 2021, Nov 17. PMID:34789874 doi:http://dx.doi.org/10.1038/s41586-021-04126-6
- ↑ 4.0 4.1 4.2 4.3 4.4 Yang F, Guo L, Li Y, Wang G, Wang J, Zhang C, Fang GX, Chen X, Liu L, Yan X, Liu Q, Qu C, Xu Y, Xiao P, Zhu Z, Li Z, Zhou J, Yu X, Gao N, Sun JP. Structure, function and pharmacology of human itch receptor complexes. Nature. 2021 Dec;600(7887):164-169. doi: 10.1038/s41586-021-04077-y. Epub 2021, Nov 17. PMID:34789875 doi:http://dx.doi.org/10.1038/s41586-021-04077-y
- ↑ Kamato D, Thach L, Bernard R, Chan V, Zheng W, Kaur H, Brimble M, Osman N, Little PJ. Structure, Function, Pharmacology, and Therapeutic Potential of the G Protein, Galpha/q,11. Front Cardiovasc Med. 2015 Mar 24;2:14. doi: 10.3389/fcvm.2015.00014. eCollection, 2015. PMID:26664886 doi:http://dx.doi.org/10.3389/fcvm.2015.00014
- ↑ Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090-109. doi: 10.2174/092986712799320556. PMID:22300046 doi:http://dx.doi.org/10.2174/092986712799320556