This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1723
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
=== MRGPRs === | === MRGPRs === | ||
| - | |||
| - | Structure with <scene name='90/904327/Overview_x2_c/1'>cortistatin</scene> | ||
| - | Structure with <scene name='90/904328/Zinc/1'>zinc</scene> | ||
The human itch GPCR, or Mas-related G-protein coupled receptor (MRGPR), is a Class A GPCR found in human sensory neurons and is responsible for the sensation of “itching” caused by skin irritation and diseases, insect bites, and hypersensitivity to certain drugs. There are currently four groups consisting of MRGPRX1, MRGPRX2, MRGPRX3, and MRGPRX4. In particular, MRGPRX4 is responsible for cholestatic itch while MRGPRX2 regulates degranulation and hypersensitivity itch reactions <ref name="Cao">Cao, Can, et al. "Structure, function and pharmacology of human itch GPCRs." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04126-6</ref>. These two, chiefly MRGPRX2, are often targets for drugs that result in mast cell degranulation and hypersensitivity side effects. In comparison to other Class A GPCRs, MRGPRX2 binds to an even wider range of ligands, including agonists such as cations and peptides. | The human itch GPCR, or Mas-related G-protein coupled receptor (MRGPR), is a Class A GPCR found in human sensory neurons and is responsible for the sensation of “itching” caused by skin irritation and diseases, insect bites, and hypersensitivity to certain drugs. There are currently four groups consisting of MRGPRX1, MRGPRX2, MRGPRX3, and MRGPRX4. In particular, MRGPRX4 is responsible for cholestatic itch while MRGPRX2 regulates degranulation and hypersensitivity itch reactions <ref name="Cao">Cao, Can, et al. "Structure, function and pharmacology of human itch GPCRs." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04126-6</ref>. These two, chiefly MRGPRX2, are often targets for drugs that result in mast cell degranulation and hypersensitivity side effects. In comparison to other Class A GPCRs, MRGPRX2 binds to an even wider range of ligands, including agonists such as cations and peptides. | ||
| Line 25: | Line 22: | ||
<scene name='90/904328/Ziz/1'>ZIZ</scene> | <scene name='90/904328/Ziz/1'>ZIZ</scene> | ||
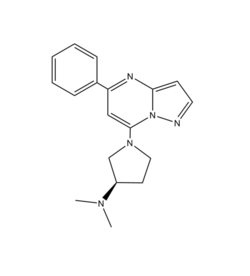
| - | (R)- | + | <scene name='90/904328/Zinc/1'>(R)-Zinc-3573</scene>is a cation ligand that binds to MRGPRX2. Its N-dimethyl is inserted into a cavity of aromatic amino acid residues Phe-170, Trp-243, and Phe-244. In this cavity, it makes ion pairs with Asp-184 and Glu-164. It is then stabilized by stacking its ring with Trp-248 and the Cys-168 to Cys-180 disulfide bond <ref name="Yang">Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y</ref>. |
[[Image: Zinc.PNG|250px|left|thumb|'''Figure X''': Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.]] | [[Image: Zinc.PNG|250px|left|thumb|'''Figure X''': Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.]] | ||
| Line 33: | Line 30: | ||
<scene name='90/904327/Zic14/1'>ZIC</scene> | <scene name='90/904327/Zic14/1'>ZIC</scene> | ||
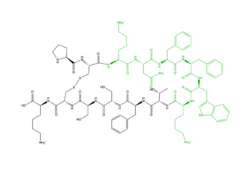
| - | Cortistatin-14 | + | <scene name='90/904327/Overview_x2_c/1'>Cortistatin-14</scene> is one of the peptide ligands that binds to MRGPRX2. Cortistatin-14 interacts with the binding pocket through an electrostatic (ZIZ) interaction in sub-pocket 1 between Lys-3 on the peptide and Glu-164 and Asp-184 on MRGPRX2 <ref name="Cao"/>. Additionally, there are hydrophobic interactions in sub-pocket 2 between the peptide and the binding pocket due to the large hydrophobic amino acids on Cortistatin-14. |
[[Image: Peptide.PNG|250px|right|thumb|'''Figure X''': Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.]] | [[Image: Peptide.PNG|250px|right|thumb|'''Figure X''': Snake Plot of GCGR TMD. Residues of particular importance in glucagon binding affinity are found in green, yellow, and black. Residues in red are the location of critical disulfide bonds, while blue residues were found to be highly conserved across all class B GPCRs.]] | ||
Revision as of 18:38, 30 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Itch GPCR
| |||||||||||
References
- ↑ Thal, David M., et al. "Structural insights into G-protein-coupled receptor allostery." Nature, Nature Publishing Group, 04 July 2018, https://www.nature.com/articles/s41586-018-0259-z
- ↑ Zhang D, Zhao Q, Wu B. Structural Studies of G Protein-Coupled Receptors. Mol Cells. 2015 Oct;38(10):836-42. doi: 10.14348/molcells.2015.0263. Epub 2015, Oct 15. PMID:26467290 doi:http://dx.doi.org/10.14348/molcells.2015.0263
- ↑ 3.0 3.1 Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, Cai X, Dai A, Jang W, Shakhnovich EI, Liu ZJ, Stevens RC, Lambert NA, Babu MM, Wang MW, Zhao S. Common activation mechanism of class A GPCRs. Elife. 2019 Dec 19;8. pii: 50279. doi: 10.7554/eLife.50279. PMID:31855179 doi:http://dx.doi.org/10.7554/eLife.50279
- ↑ 4.0 4.1 4.2 Cao, Can, et al. "Structure, function and pharmacology of human itch GPCRs." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04126-6
- ↑ 5.0 5.1 5.2 5.3 Yang, Fan, et al. "Structure, function and pharmacology of human itch receptor complexes." Nature, Nature Publishing Group, 17 November 2021, https://www.nature.com/articles/s41586-021-04077-y
- ↑ 6.0 6.1 Schonegge, Anne-Marie, et al. "Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity." Nature, Nature Publishing Group, 18 December 2017, https://www.nature.com/articles/s41467-017-02257-x
- ↑ Katritch V, Fenalti G, Abola EE, Roth BL, Cherezov V, Stevens RC. Allosteric sodium in class A GPCR signaling. Trends Biochem Sci. 2014 May;39(5):233-44. doi: 10.1016/j.tibs.2014.03.002. Epub , 2014 Apr 21. PMID:24767681 doi:http://dx.doi.org/10.1016/j.tibs.2014.03.002