User:George G. Papadeas/Sandbox VKOR
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
=== Author's Notes === | === Author's Notes === | ||
| - | Crystal structures of Human VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA). VKA substrates utilized were anticoagulants, namely [https://en.wikipedia.org/wiki/Warfarin Warfarin], [https://en.wikipedia.org/wiki/Brodifacoum Brodifacoum], [https://en.wikipedia.org/wiki/Phenindione Phenindione], and [https://en.wikipedia.org/wiki/Chlorophacinone Chlorophacinone]. Second, VKOR-like homologs, | + | Crystal structures of Human VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA). VKA substrates utilized were anticoagulants, namely [https://en.wikipedia.org/wiki/Warfarin Warfarin], [https://en.wikipedia.org/wiki/Brodifacoum Brodifacoum], [https://en.wikipedia.org/wiki/Phenindione Phenindione], and [https://en.wikipedia.org/wiki/Chlorophacinone Chlorophacinone]. Second, VKOR-like homologs, isolated from the pufferfish ''Takifugu rubripes'', aided in structure classification as well. |
== Structural Highlights== | == Structural Highlights== | ||
| Line 23: | Line 23: | ||
== Function: Method of Coagulation == | == Function: Method of Coagulation == | ||
=== Brief Overview === | === Brief Overview === | ||
| - | The <scene name='90/906893/Open_conformation/1'>open conformation</scene> will be | + | The <scene name='90/906893/Open_conformation/1'>open conformation</scene> will be prepared for a substrate. |
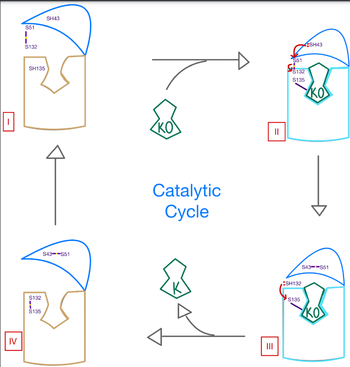
=== Catalytic Mechanism === | === Catalytic Mechanism === | ||
Revision as of 14:38, 5 April 2022
VKOR
| |||||||||||
References
1. Li, Weikai et al. “Structure of a bacterial homologue of vitamin K epoxide reductase.” Nature vol. 463,7280 (2010): 507-12. doi:10.1038/nature08720.
2. Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2021 Jan 1;371(6524):eabc5667. doi: 10.1126/science.abc5667. Epub 2020 Nov 5. PMID: 33154105; PMCID: PMC7946407.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644