Sandbox Reserved 1719
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | ==Human Itch G-Coupled Protein Receptors== | + | == Human Itch G-Coupled Protein Receptors == |
<StructureSection load='7s8l' size='350' side='right' caption='Cryo-EM structure of Gq coupled MRGPRX2.' scene='90/904324/Mrgprx2/2'> | <StructureSection load='7s8l' size='350' side='right' caption='Cryo-EM structure of Gq coupled MRGPRX2.' scene='90/904324/Mrgprx2/2'> | ||
| - | =Introduction= | + | = Introduction = |
[https://proteopedia.org/wiki/index.php/G_protein-coupled_receptors G protein-coupled receptors] (GPCRs) are the largest class of integral membrane proteins.<ref name="Zhang 2015">DOI 10.14348/molcells.2015.0263</ref> GPCRs<ref name= "Zhang 2015"/><ref>PMID: 20019124</ref> are divided into five families; the [https://proteopedia.org/wiki/index.php/Sandbox_Reserved_895 rhodopsin family (class A)], the [https://proteopedia.org/wiki/index.php/4ers secretin family (class B)], the [https://proteopedia.org/wiki/index.php/6wiv glutamate family (class C)], the [https://proteopedia.org/wiki/index.php/6bd4 frizzled/taste family (class F)], and the [https://en.wikipedia.org/wiki/Adhesion_G_protein-coupled_receptor adhesion family].<ref name= "Zhang 2015"/><ref name= "Zhang 2006"/> All GPCRs contain a similar seven α-helical transmembrane domain (TMD) that undergoes a conformation change once bound to its ligand. This conformational change then transduces a signal to a coupled, heterotrimeric G protein which then dictates whether an intracellular signaling pathway will be initiated or inhibited. The initiation of the intracellular signaling pathway occurs in response to a variety of stimuli such as light, Ca<sup>2+</sup>, peptides, different proteins, and many more stimuli. Ultimately, intracellular signaling [https://en.wikipedia.org/wiki/G_protein%E2%80%93coupled_receptor#Physiological_roles accomplishes many interesting physiological roles].<ref name= "Zhang 2015"/><ref name= "Zhang 2006">DOI 10.1371/journal.pcbi.0020013</ref> | [https://proteopedia.org/wiki/index.php/G_protein-coupled_receptors G protein-coupled receptors] (GPCRs) are the largest class of integral membrane proteins.<ref name="Zhang 2015">DOI 10.14348/molcells.2015.0263</ref> GPCRs<ref name= "Zhang 2015"/><ref>PMID: 20019124</ref> are divided into five families; the [https://proteopedia.org/wiki/index.php/Sandbox_Reserved_895 rhodopsin family (class A)], the [https://proteopedia.org/wiki/index.php/4ers secretin family (class B)], the [https://proteopedia.org/wiki/index.php/6wiv glutamate family (class C)], the [https://proteopedia.org/wiki/index.php/6bd4 frizzled/taste family (class F)], and the [https://en.wikipedia.org/wiki/Adhesion_G_protein-coupled_receptor adhesion family].<ref name= "Zhang 2015"/><ref name= "Zhang 2006"/> All GPCRs contain a similar seven α-helical transmembrane domain (TMD) that undergoes a conformation change once bound to its ligand. This conformational change then transduces a signal to a coupled, heterotrimeric G protein which then dictates whether an intracellular signaling pathway will be initiated or inhibited. The initiation of the intracellular signaling pathway occurs in response to a variety of stimuli such as light, Ca<sup>2+</sup>, peptides, different proteins, and many more stimuli. Ultimately, intracellular signaling [https://en.wikipedia.org/wiki/G_protein%E2%80%93coupled_receptor#Physiological_roles accomplishes many interesting physiological roles].<ref name= "Zhang 2015"/><ref name= "Zhang 2006">DOI 10.1371/journal.pcbi.0020013</ref> | ||
| Line 24: | Line 24: | ||
<scene name='90/904324/7s8p_mx4/4'>MRGPRX4</scene> is another sub-group of the MRGPRX family, which mediates cholestatic itch and is a target of nateglinide drugs.<ref name="Can"/><ref name="Yang"/> MRGPRX4 also couples to G<sub>q</sub> and G<sub>i</sub> similar to MRGPRX2. | <scene name='90/904324/7s8p_mx4/4'>MRGPRX4</scene> is another sub-group of the MRGPRX family, which mediates cholestatic itch and is a target of nateglinide drugs.<ref name="Can"/><ref name="Yang"/> MRGPRX4 also couples to G<sub>q</sub> and G<sub>i</sub> similar to MRGPRX2. | ||
| - | =Structure Overview= | + | = Structure Overview = |
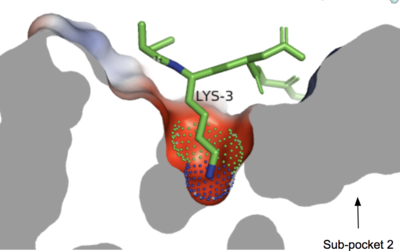
The <scene name='90/904324/Mrgprx2/5'>MRGPRX2</scene> receptor <scene name='90/904324/Structure_overview_of_tmecicl/2'>contains 7 transmembrane helices (TM 1-7), 3 intracellular loops (ICL 1-3), and 3 extracellular loops (ECL 1-3).</scene> Ligand binding occurs at the N-terminus in the extracellular domain and is composed of two binding sub-pockets.<ref name="Can"/> The intracellular domain consists of helix VII and a C-terminal sequence, which binds the G-protein and promotes downstream signaling.<ref name="Can"/> | The <scene name='90/904324/Mrgprx2/5'>MRGPRX2</scene> receptor <scene name='90/904324/Structure_overview_of_tmecicl/2'>contains 7 transmembrane helices (TM 1-7), 3 intracellular loops (ICL 1-3), and 3 extracellular loops (ECL 1-3).</scene> Ligand binding occurs at the N-terminus in the extracellular domain and is composed of two binding sub-pockets.<ref name="Can"/> The intracellular domain consists of helix VII and a C-terminal sequence, which binds the G-protein and promotes downstream signaling.<ref name="Can"/> | ||
[[Image:Structure_overview_of_Mx_with_membrane.PNG|500px|center|thumb|'''Figure 1.''' Location of MRGPRX2 in plasma membrane.]] | [[Image:Structure_overview_of_Mx_with_membrane.PNG|500px|center|thumb|'''Figure 1.''' Location of MRGPRX2 in plasma membrane.]] | ||
| Line 83: | Line 83: | ||
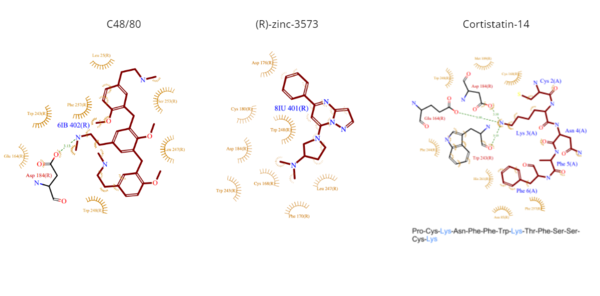
[[Image:Ligands3.png|600px|center|thumb|'''Figure 5.''' Common MRGPRX2 ligand structures shown in brown. Hydrophobic interactions shown by dashed wheat lines indicating direction. Positive atoms are represented in blue. Negative atoms are represented in red.]] | [[Image:Ligands3.png|600px|center|thumb|'''Figure 5.''' Common MRGPRX2 ligand structures shown in brown. Hydrophobic interactions shown by dashed wheat lines indicating direction. Positive atoms are represented in blue. Negative atoms are represented in red.]] | ||
| - | == Differences | + | == Differences to most class A GPCRs == |
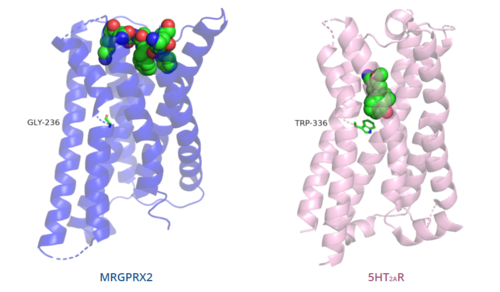
The unique characteristics of <scene name='90/904324/Mrgprx2/5'>MRGPRX2</scene> in comparison to class A GPCRs provides an explanation for the differences in ligand interactions. These differences in intermolecular interactions and structural motifs contribute to the surface level ligand binding in MRGPRX2, whereas the typical ligand interaction occurs deep within the helices in class A GPCRs. | The unique characteristics of <scene name='90/904324/Mrgprx2/5'>MRGPRX2</scene> in comparison to class A GPCRs provides an explanation for the differences in ligand interactions. These differences in intermolecular interactions and structural motifs contribute to the surface level ligand binding in MRGPRX2, whereas the typical ligand interaction occurs deep within the helices in class A GPCRs. | ||
[[Image:Comparison_of_binding_depth.PNG|500px|center|thumb|'''Figure 6.''' Comparison of ligand binding depth in MRGPRX2 (blue) and 5HT2AR (purple).]] | [[Image:Comparison_of_binding_depth.PNG|500px|center|thumb|'''Figure 6.''' Comparison of ligand binding depth in MRGPRX2 (blue) and 5HT2AR (purple).]] | ||
| - | ===Toggle | + | === Toggle switch === |
The conserved <scene name='90/904324/5ht2a_toggle_switch/3'>toggle switch</scene> of class A GPCRs enables the receptor to initiate the signaling cascade. However, MRGPRX2 does not contain the conserved ‘toggle switch’ Trp. Instead, it is replaced by <scene name='90/904324/Toggle_switch/7'>Gly</scene>. Therefore, the main residues of this motif in MRGPRX2 are Gly236, Tyr113, Phe239, and Trp243.<ref name="Can"/> As a result, TM6 is shifted closer to TM3 on the extracellular side of the membrane. This conformational change may account for the lack of ligand binding of MRGPRX2 as compared to family A receptors.<ref name="Can"/> This toggle switch swap also means that ligands, such as (R)-zinc-3573 and Cortistatin-14, bind in a different spot than ligands do on other class A GPCRs. In MRGPRX2, Gly236 is located closer to the bottom of the interface, which is the same in MRGPRX4 (Gly229). Compared with other structures, such as 5-HT<sub>2A</sub>R (PDB ID 6WHA), A<sub>2A</sub>R (PDB ID 5G53), and β<sub>2</sub>AR (PDB ID 3SN6), the TM6 helices of MRGPRX2 and MRGPRX4 are closer to the TM3 helix which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | The conserved <scene name='90/904324/5ht2a_toggle_switch/3'>toggle switch</scene> of class A GPCRs enables the receptor to initiate the signaling cascade. However, MRGPRX2 does not contain the conserved ‘toggle switch’ Trp. Instead, it is replaced by <scene name='90/904324/Toggle_switch/7'>Gly</scene>. Therefore, the main residues of this motif in MRGPRX2 are Gly236, Tyr113, Phe239, and Trp243.<ref name="Can"/> As a result, TM6 is shifted closer to TM3 on the extracellular side of the membrane. This conformational change may account for the lack of ligand binding of MRGPRX2 as compared to family A receptors.<ref name="Can"/> This toggle switch swap also means that ligands, such as (R)-zinc-3573 and Cortistatin-14, bind in a different spot than ligands do on other class A GPCRs. In MRGPRX2, Gly236 is located closer to the bottom of the interface, which is the same in MRGPRX4 (Gly229). Compared with other structures, such as 5-HT<sub>2A</sub>R (PDB ID 6WHA), A<sub>2A</sub>R (PDB ID 5G53), and β<sub>2</sub>AR (PDB ID 3SN6), the TM6 helices of MRGPRX2 and MRGPRX4 are closer to the TM3 helix which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | ||
| - | ===PIF/LLF motif=== | + | === PIF/LLF motif === |
The MRGPRX2 structure does not contain the conserved <scene name='90/904324/5ht2a/3'>PIF motif</scene> at the TM3-TM6 interface.<ref name="Can"/> Canonically, the PIF motif consists of a Pro, Ile, and Phe which transduce the signal produce by ligand binding through the TMD within conserved distances (5.50Å, 3.40Å, and 6.44Å respectively).<ref name="Can"/><ref>DOI: 10.1038/s41467-017-02257-x</ref> In this motif, the residues are not conserved at specific positions in the amino acid sequence, but instead are conserved at distances that allow them to interact.<ref name="Can"/> | The MRGPRX2 structure does not contain the conserved <scene name='90/904324/5ht2a/3'>PIF motif</scene> at the TM3-TM6 interface.<ref name="Can"/> Canonically, the PIF motif consists of a Pro, Ile, and Phe which transduce the signal produce by ligand binding through the TMD within conserved distances (5.50Å, 3.40Å, and 6.44Å respectively).<ref name="Can"/><ref>DOI: 10.1038/s41467-017-02257-x</ref> In this motif, the residues are not conserved at specific positions in the amino acid sequence, but instead are conserved at distances that allow them to interact.<ref name="Can"/> | ||
In the MRGPRX2 <scene name='90/904324/Pifllf_motif/5'>LLF motif</scene>, the residues that make up TM5 have shifted down two residues making Leu194 analogous to the position of the Pro in other GPCRs. However, in MRGPRX2, Leu194 is slightly closer to the other residues in the motif at 5.48Å.<ref name="Can"/> The residue at a distance of 5.50Å in MRGPRX2 is Met196. It does not interact with the motif because it is angled away from the TM3 and TM6 interface.<ref name="Can"/> Leu194 interacts with two other residues, Leu117 and Phe232. The additional change from Ileto Leu is why the motif in MRGPRX2 is called the LLF motif.<ref name="Can"/> Compared with other structures, such as 5-HT<sub>2A</sub>R (PDB ID 6WHA), A<sub>2A</sub>R (PDB ID 5G53), and β<sub>2</sub>AR (PDB ID 3SN6), the TM6 helices of MRGPRX2 are closer to the TM3 helix due to the shift in residues which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | In the MRGPRX2 <scene name='90/904324/Pifllf_motif/5'>LLF motif</scene>, the residues that make up TM5 have shifted down two residues making Leu194 analogous to the position of the Pro in other GPCRs. However, in MRGPRX2, Leu194 is slightly closer to the other residues in the motif at 5.48Å.<ref name="Can"/> The residue at a distance of 5.50Å in MRGPRX2 is Met196. It does not interact with the motif because it is angled away from the TM3 and TM6 interface.<ref name="Can"/> Leu194 interacts with two other residues, Leu117 and Phe232. The additional change from Ileto Leu is why the motif in MRGPRX2 is called the LLF motif.<ref name="Can"/> Compared with other structures, such as 5-HT<sub>2A</sub>R (PDB ID 6WHA), A<sub>2A</sub>R (PDB ID 5G53), and β<sub>2</sub>AR (PDB ID 3SN6), the TM6 helices of MRGPRX2 are closer to the TM3 helix due to the shift in residues which makes the binding pocket more occluded than seen in canonical structures.<ref name="Can"/> | ||
| - | ===DRY/ERC motif=== | + | === DRY/ERC motif === |
The majority of class A GPCRs have a [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY motif]. However, MRGPRX2 contains an <scene name='90/904324/Erc_motif/3'>ERC motif</scene>, which replaces Tyr174 with Cys128.<ref name="Can"/> This replacement alters the spatial organization of the helices due to the fact that Tyr is no longer present to push the helices outward.<ref name="Can"/> As a result, the binding site in MRGPRX2 is shallower which lead to surface-level ligand interactions.<ref name="Can"/> | The majority of class A GPCRs have a [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY motif]. However, MRGPRX2 contains an <scene name='90/904324/Erc_motif/3'>ERC motif</scene>, which replaces Tyr174 with Cys128.<ref name="Can"/> This replacement alters the spatial organization of the helices due to the fact that Tyr is no longer present to push the helices outward.<ref name="Can"/> As a result, the binding site in MRGPRX2 is shallower which lead to surface-level ligand interactions.<ref name="Can"/> | ||
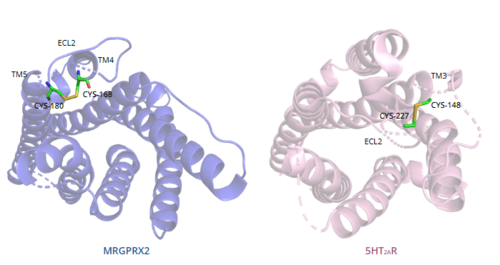
| - | ===Disulfide bonds=== | + | === Disulfide bonds === |
In general, class A GPCRs have a conserved disulfide bond between TM3 and ECL2.<ref name="Can"/> In contrast, the MRGPRX2 <scene name='90/904324/Mrgprx2_disulfide_bonds/4'>disulfide bond</scene> is located between Cys168 of TM4 and Cys180 of TM5, which structurally flips ECL2 to the top of TM4 and TM5.<ref name="Can"/> This creates the wide ligand-binding surface of MRGPRX2 that contributes to surface level binding which allows diverse ligand interactions.<ref name="Can"/> | In general, class A GPCRs have a conserved disulfide bond between TM3 and ECL2.<ref name="Can"/> In contrast, the MRGPRX2 <scene name='90/904324/Mrgprx2_disulfide_bonds/4'>disulfide bond</scene> is located between Cys168 of TM4 and Cys180 of TM5, which structurally flips ECL2 to the top of TM4 and TM5.<ref name="Can"/> This creates the wide ligand-binding surface of MRGPRX2 that contributes to surface level binding which allows diverse ligand interactions.<ref name="Can"/> | ||
[[Image:Disulfide_bond_comparison.png|500px|center|thumb|'''Figure 7.''' Comparison of disulfide bond location in MRGPRX2 (blue) and 5HT2AR (purple).]] | [[Image:Disulfide_bond_comparison.png|500px|center|thumb|'''Figure 7.''' Comparison of disulfide bond location in MRGPRX2 (blue) and 5HT2AR (purple).]] | ||
| - | ===Sodium binding site=== | + | === Sodium binding site === |
The <scene name='90/904324/Mrgprx2_sodium_site/2'>sodium binding site</scene> of MRGPRX2 contains the conserved Asp75, however, the conserved Ser116 on TM3 seen in class A GPCRs is replaced by Gly116.<ref name="Can"/> The presence of Gly as opposed to the polar Ser contributes to a lack of polar character in addition to decreasing the size of the binding pocket. The sodium pocket generally serves as an allosteric binding pocket for sodium. Both factors thereby limit the binding of sodium ions to the [https://proteopedia.org/wiki/index.php/Neurotensin_receptor#sodium%20binding%20pocket sodium binding site] in MRGPRX2. | The <scene name='90/904324/Mrgprx2_sodium_site/2'>sodium binding site</scene> of MRGPRX2 contains the conserved Asp75, however, the conserved Ser116 on TM3 seen in class A GPCRs is replaced by Gly116.<ref name="Can"/> The presence of Gly as opposed to the polar Ser contributes to a lack of polar character in addition to decreasing the size of the binding pocket. The sodium pocket generally serves as an allosteric binding pocket for sodium. Both factors thereby limit the binding of sodium ions to the [https://proteopedia.org/wiki/index.php/Neurotensin_receptor#sodium%20binding%20pocket sodium binding site] in MRGPRX2. | ||
| - | =Mechanism= | + | = Mechanism = |
Binding to the extracellular N-terminus domain triggers a transmembrane conformation change of MRGPRX2, which demonstrates a less significant change when compared to other class A GPCRs due to the surface level binding of the ligand to MRGPRX2.<ref name="Can"/> Once ligand binding and the conformational change to the active state have taken place, the signal is relayed to the α-subunit of the heterotrimeric G-protein.<ref name="nelson"/> The α-subunit will then exchange a GDP for GTP to initiate the dissociation of the α, β, and γ subunits.<ref name="nelson"/> During this dissociation, the α-subunit is able to travel away from the receptor in the plane of the membrane to bind to downstream effectors to produce a cellular response.<ref name="nelson"/> | Binding to the extracellular N-terminus domain triggers a transmembrane conformation change of MRGPRX2, which demonstrates a less significant change when compared to other class A GPCRs due to the surface level binding of the ligand to MRGPRX2.<ref name="Can"/> Once ligand binding and the conformational change to the active state have taken place, the signal is relayed to the α-subunit of the heterotrimeric G-protein.<ref name="nelson"/> The α-subunit will then exchange a GDP for GTP to initiate the dissociation of the α, β, and γ subunits.<ref name="nelson"/> During this dissociation, the α-subunit is able to travel away from the receptor in the plane of the membrane to bind to downstream effectors to produce a cellular response.<ref name="nelson"/> | ||
Revision as of 23:21, 17 April 2022
Human Itch G-Coupled Protein Receptors
| |||||||||||
Student contributors
Madeline Beck
Joey Gareis