User:George G. Papadeas/Sandbox VKOR
From Proteopedia
(Difference between revisions)
| Line 32: | Line 32: | ||
== Disease and Treatment == | == Disease and Treatment == | ||
=== Afflictions === | === Afflictions === | ||
| - | + | Since activated Vitamin K plays a crucial role in blood coagulation, defects in the function and enzymatic activity of VKOR may detrimentally effect on Vitamin K's ability to promote blood clotting. Mutations in VKOR also increase susceptibility to vascular diseases, such as a stroke [https://doi.org/10.1161/CIRCULATIONAHA.105.580167]. Vitamin K is also important in maintaining bone health with inactivity of VKOR linked to decreased bone density and osteoporosis [https://doi.org/10.7759/cureus.10816]. | |
=== Inhibition === | === Inhibition === | ||
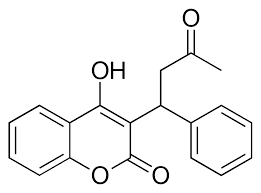
| - | [[Image:Warfarin.png |400 px| right| thumb]] | + | [[Image:Warfarin.png |400 px| right| thumb | Figure 4. Structure of Warfarin.]] |
| - | The most inexpensive and common way to treat blood clotting is through the VKOR inhibitor, <scene name='90/906893/Vkor_with_warfarin_bound/2'>Warfarin</scene>. [https://en.wikipedia.org/wiki/Warfarin Warfarin] is able to do so by outcompeting KO. | + | The most inexpensive and common way to treat blood clotting is through the VKOR inhibitor, <scene name='90/906893/Vkor_with_warfarin_bound/2'>Warfarin</scene>. [https://en.wikipedia.org/wiki/Warfarin Warfarin] is able to do so by outcompeting KO, such that Vitamin K cannot be activated to promote coagulation in the blood. Warfarin will enter the binding pocket of VKOR, creating strong hydrogen bonds with the active site. Warfarin resistance may also occur due to mutations of VKOR, decreasing the effective anticoagulation some drugs may be attempting to promote. The degree of resistance is important to determine so that warfarin may be an effective anticoagulant without being detrimentally effective in blood flow. |
=== Mutations === | === Mutations === | ||
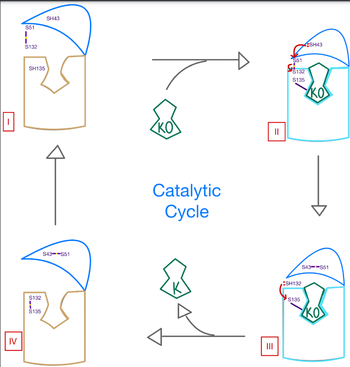
Some key <scene name='90/906893/Active_site_mutations/2'>mutations</scene> that can be detrimental to the VKOR structure are mutations of the <scene name='90/906893/Active_site/4'>active site</scene>. The two main residues, N80 and Y139, can be mutated to A80 and F139 creating a decrease in recognition and stabilization. | Some key <scene name='90/906893/Active_site_mutations/2'>mutations</scene> that can be detrimental to the VKOR structure are mutations of the <scene name='90/906893/Active_site/4'>active site</scene>. The two main residues, N80 and Y139, can be mutated to A80 and F139 creating a decrease in recognition and stabilization. | ||
| + | |||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 02:57, 18 April 2022
VKOR
| |||||||||||
References
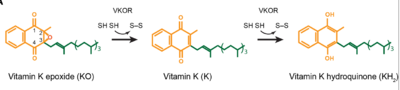
1. DJin, Da-Yun, Tie, Jian-Ke, and Stafford, Darrel W. "The Conversion of Vitamin K Epoxide to Vitamin K Quinone and Vitamin K Quinone to Vitamin K Hydroquinone Uses the Same Active Site Cysteines." Biochemistry 2007 46 (24), 7279-7283 [1].
2. Li, Weikai et al. “Structure of a bacterial homologue of vitamin K epoxide reductase.” Nature vol. 463,7280 (2010): 507-12. doi:10.1038/nature08720.
3. Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2021 Jan 1;371(6524):eabc5667. doi: 10.1126/science.abc5667. Epub 2020 Nov 5. PMID: 33154105; PMCID: PMC7946407.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Unknown PubmedID 10.1126
- ↑ Unknown PubmedID 10.1021
- ↑ Unknown PubmedID 10.1126