We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1719
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
[[Image:Electro_map_with_cortistatin.png|400px|center|thumb|'''Figure 2.''' Electrostatic surface of MRGPRX2 ligand binding pocket with cortistatin-14]] | [[Image:Electro_map_with_cortistatin.png|400px|center|thumb|'''Figure 2.''' Electrostatic surface of MRGPRX2 ligand binding pocket with cortistatin-14]] | ||
| - | [[Image:Subpocket1.jpg.png|200px|left|thumb|'''Figure 3.''' Cross-sectional view of electrostatic surface of MRGPRX2 sub-pocket 1 interaction with lysine 3 of cortistatin-14.]][[Image:Screen Shot 2022-03-27 at 3.40.41 PM.png|200px|left|thumb|'''Figure 4.''' Cross-sectional view of electrostatic surface of MRGPRX2 sub-pocket 2.]] | ||
==== Sub-pocket 1 ==== | ==== Sub-pocket 1 ==== | ||
| - | <scene name='90/904324/Active_site_residues/9'>Sub-pocket 1</scene> is a small, shallow pocket formed by TM3, TM6, and ECL2.<ref name="Can"/> Ligand binding is mediated by two key residues on the MRGPRX2 protein within the binding site: Glu164 and Asp184.<ref name="Can"/> The strong charge interactions of these two residues create a highly negatively electrostatic interaction crucial for binding cationic ligands, namely those containing arginine or lysine as shown in '''Figure | + | <scene name='90/904324/Active_site_residues/9'>Sub-pocket 1</scene> is a small, shallow pocket formed by TM3, TM6, and ECL2.<ref name="Can"/> Ligand binding is mediated by two key residues on the MRGPRX2 protein within the binding site: Glu164 and Asp184.<ref name="Can"/> The strong charge interactions of these two residues create a highly negatively electrostatic interaction crucial for binding cationic ligands, namely those containing arginine or lysine as shown in '''Figure 3A'''.<ref name="Can"/>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| Line 49: | Line 42: | ||
==== Sub-pocket 2 ==== | ==== Sub-pocket 2 ==== | ||
| - | <scene name='90/904324/Active_site_residues/8'>Sub-pocket 2</scene> is formed by TM1, TM2, TM6, and TM7.<ref name="Can"/> This binding sub-pocket is much broader and allows for the binding of larger structures ('''Figure | + | <scene name='90/904324/Active_site_residues/8'>Sub-pocket 2</scene> is formed by TM1, TM2, TM6, and TM7.<ref name="Can"/> This binding sub-pocket is much broader and allows for the binding of larger structures ('''Figure 3B''').<ref name="Can"/> The key residues involved are Trp243 and Phe170 that contributes to the high hydrophobicity of the binding pocket.<ref name="Can"/> The hydrophobicity of this binding pocket accounts for the large electrostatic difference observed between the two sub pockets demonstrated in '''Figure 2'''. |
| - | + | ||
| - | + | ||
| + | [[Image:Screen Shot 2022-04-18 at 1.35.31 PM.png|400px|center|thumb|'''Figure 3.''' (A). Cross-sectional views of electrostatic surface of MRGPRX2 sub-pocket 1 interaction with lysine 3 and (B) sub-pocket 2 interaction with phenylalanine 6 of cortistatin-14.] | ||
| Line 67: | Line 59: | ||
*<scene name='90/904324/Cortistatin-14/6'>Cortistatin-14</scene> | *<scene name='90/904324/Cortistatin-14/6'>Cortistatin-14</scene> | ||
**Cortistatin-14 is an endogenous, cyclic, neuropeptide agonist which interacts with MRGPRX2 in the same way whether it is coupled to G<sub>i</sub> or G<sub>q</sub> proteins.<ref name="Can"/><ref name= "Jiang">DOI: 10.3389/fphar.2018.00767</ref> Cortistation-14 is widely available in many systems throughout the body and naturally functions to regulate many physiological and pathological mechanisms. The lysine residue (Lys3) on Cortistatin-14 binds in the negatively-charged sub-pocket 1 and forms strong charge interactions with Asp184 and Glu164.<ref name="Can"/> The remaining residues of Cortistatin-14 will extend over to sub-pocket 2 and bind through hydrophobic interactions.<ref name="Can"/> | **Cortistatin-14 is an endogenous, cyclic, neuropeptide agonist which interacts with MRGPRX2 in the same way whether it is coupled to G<sub>i</sub> or G<sub>q</sub> proteins.<ref name="Can"/><ref name= "Jiang">DOI: 10.3389/fphar.2018.00767</ref> Cortistation-14 is widely available in many systems throughout the body and naturally functions to regulate many physiological and pathological mechanisms. The lysine residue (Lys3) on Cortistatin-14 binds in the negatively-charged sub-pocket 1 and forms strong charge interactions with Asp184 and Glu164.<ref name="Can"/> The remaining residues of Cortistatin-14 will extend over to sub-pocket 2 and bind through hydrophobic interactions.<ref name="Can"/> | ||
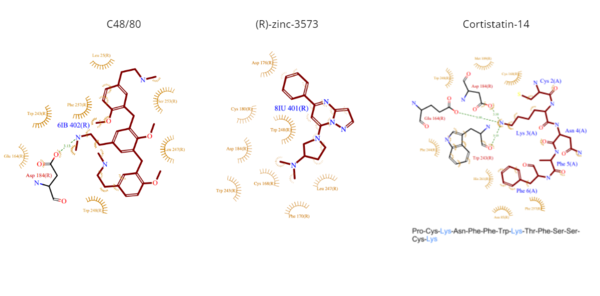
| - | [[Image:Ligands3.png|600px|center|thumb|'''Figure | + | [[Image:Ligands3.png|600px|center|thumb|'''Figure 4.''' Common MRGPRX2 ligand structures and interactions. Hydrophobic interactions shown by dashed wheat lines indicating direction. Positive atoms are represented in blue. Negative atoms are represented in red.]] |
== Differences to most class A GPCRs == | == Differences to most class A GPCRs == | ||
| Line 78: | Line 70: | ||
</jmol> | </jmol> | ||
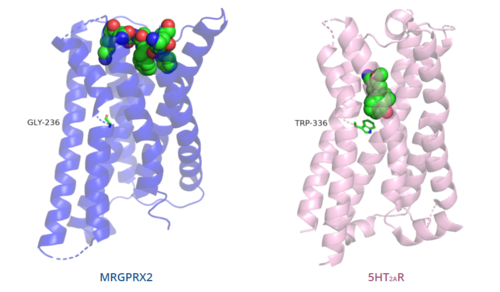
| - | [[Image:Comparison_of_binding_depth.PNG|500px|center|thumb|'''Figure | + | [[Image:Comparison_of_binding_depth.PNG|500px|center|thumb|'''Figure 5.''' Comparison of ligand binding depth in MRGPRX2 (blue) and 5HT2AR (purple).]] |
=== Toggle switch === | === Toggle switch === | ||
| - | The conserved <scene name='90/904324/5ht2a_toggle_switch/3'>toggle switch</scene> of class A GPCRs acts to activate or inhibit the transduction of the signaling cascade. Typical class A GPCRs contain the conserved ‘toggle switch’ Trp336. In MRGPRX2, this residue is replaced by <scene name='90/904324/Toggle_switch/7'>Gly236</scene>. <ref name="Can"/> By substituting the larger Trp residue with a smaller Gly residue, TM6 is shifted closer to TM3 on the extracellular side of the membrane and contributes to the more tightly packed and shallow binding pocket as compared to other canonical structures. The occluded binding pocket contributes the surface level binding depicted in '''Figure | + | The conserved <scene name='90/904324/5ht2a_toggle_switch/3'>toggle switch</scene> of class A GPCRs acts to activate or inhibit the transduction of the signaling cascade. Typical class A GPCRs contain the conserved ‘toggle switch’ Trp336. In MRGPRX2, this residue is replaced by <scene name='90/904324/Toggle_switch/7'>Gly236</scene>. <ref name="Can"/> By substituting the larger Trp residue with a smaller Gly residue, TM6 is shifted closer to TM3 on the extracellular side of the membrane and contributes to the more tightly packed and shallow binding pocket as compared to other canonical structures. The occluded binding pocket contributes the surface level binding depicted in '''Figure 5''', and allows for a greater variety of ligands interactions with MRGPRX2 as compared to other class A GPCRs, such as [https://proteopedia.org/wiki/index.php/5-hydroxytryptamine_receptor 5-HT<sub>2A</sub>R], [https://proteopedia.org/wiki/index.php/Adrenergic_receptor A<sub>2A</sub>R], and [https://proteopedia.org/wiki/index.php/Beta-2_Adrenergic_Receptor β<sub>2</sub>AR].<ref name="Can"/> |
=== PIF/LLF motif === | === PIF/LLF motif === | ||
| Line 91: | Line 83: | ||
=== DRY/ERC motif === | === DRY/ERC motif === | ||
The majority of class A GPCRs have a [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY motif] that is responsible for creating salt bridges that maintain the inactive conformation of the receptor until ligand binding.<ref name="Can"/> | The majority of class A GPCRs have a [https://proteopedia.org/wiki/index.php/A_Physical_Model_of_the_%CE%B22-Adrenergic_Receptor#conserved%20DRY%20motif conserved E/DRY motif] that is responsible for creating salt bridges that maintain the inactive conformation of the receptor until ligand binding.<ref name="Can"/> | ||
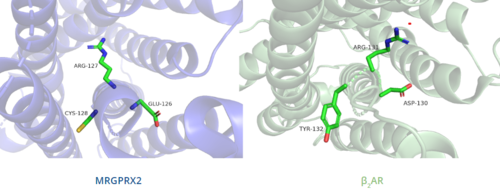
| - | In contrast, '''Figure | + | In contrast, '''Figure 6''' shows MRGPRX2 containing an <scene name='90/904324/Erc_motif/3'>ERC motif</scene> in place of the E/DRY motif, which replaces Tyr174 with Cys128.<ref name="Can"/> This replacement alters the spatial organization of the helices due to the replacement of the larger Tyr residue with the smaller Cys residue, thereby condensing the helices.<ref name="Can"/> The condensed spatial organization in MRGPRX2 accounts for the less significant change once a ligand binds to the receptor.<ref name="Can"/> |
| - | [[Image:B2AR_motif.png|500px|center|thumb|'''Figure | + | [[Image:B2AR_motif.png|500px|center|thumb|'''Figure 6.''' Comparison of ERC motif in MRGPRX2 (blue) and conserved DRY motif in β<sub>2</sub>AR (green).]] |
=== Disulfide bonds === | === Disulfide bonds === | ||
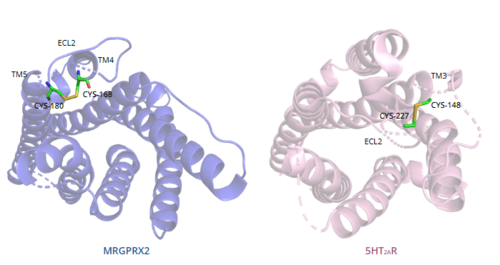
| - | In general, class A GPCRs have a <scene name='90/904324/5ht2a/4'>conserved disulfide bond</scene> between TM3 and ECL2.<ref name="Can"/> In contrast, '''Figure | + | In general, class A GPCRs have a <scene name='90/904324/5ht2a/4'>conserved disulfide bond</scene> between TM3 and ECL2.<ref name="Can"/> In contrast, '''Figure 7''' shows the location MRGPRX2 <scene name='90/904324/Mrgprx2_disulfide_bonds/5'>disulfide bond</scene> between Cys168 of TM4 and Cys180 of TM5, which structurally flips ECL2 to the top of TM4 and TM5.<ref name="Can"/> This creates the wide ligand-binding surface of MRGPRX2 that contributes to surface level binding which allows diverse ligand interactions.<ref name="Can"/> |
| - | [[Image:Disulfide_bond_comparison.png|500px|center|thumb|'''Figure | + | [[Image:Disulfide_bond_comparison.png|500px|center|thumb|'''Figure 7.''' Comparison of disulfide bond location in MRGPRX2 (blue) and 5HT2AR (purple).]] |
=== Sodium binding site === | === Sodium binding site === | ||
| Line 110: | Line 102: | ||
The conformations of the G-proteins vary based on their association with a particular membrane receptor due to interactions between the amino acids in the N-terminus of the α subunit and the C-terminus of the receptor.<ref name="Kamato"/> | The conformations of the G-proteins vary based on their association with a particular membrane receptor due to interactions between the amino acids in the N-terminus of the α subunit and the C-terminus of the receptor.<ref name="Kamato"/> | ||
| - | [[Image:Mast cell mechanism.png|400px|right|thumb|'''Figure | + | [[Image:Mast cell mechanism.png|400px|right|thumb|'''Figure 8.''' Cellular response of mast cell upon activation of MRGPRX2.]] |
Revision as of 17:57, 18 April 2022
Human Itch G-Coupled Protein Receptors
| |||||||||||