User:George G. Papadeas/Sandbox VKOR
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
=== Author's Notes === | === Author's Notes === | ||
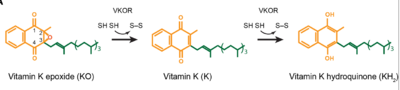
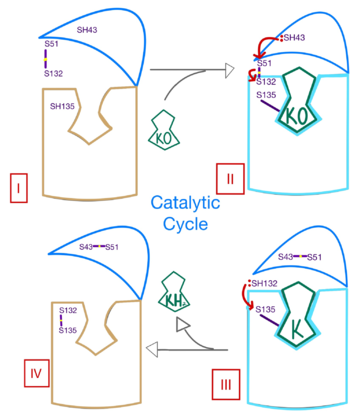
| - | Structural characterization of VKOR has been difficult due to its in vitro instability. Recently, a series of atomic structures have been determined utilizing anticoagulant stabilization and VKOR-like homologs<ref>DOI 10.1126/science.abc5667</ref>. Crystal structures of VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA)<ref>DOI 10.1126/science.abc5667</ref>. VKA substrates utilized were anticoagulants, namely [https://en.wikipedia.org/wiki/Warfarin Warfarin], [https://en.wikipedia.org/wiki/Brodifacoum Brodifacoum], [https://en.wikipedia.org/wiki/Phenindione Phenindione], and [https://en.wikipedia.org/wiki/Chlorophacinone Chlorophacinone]. Second, VKOR-like homologs were utilized to aid in structure classification. Homologs refer to specific cysteine residues that have been mutated to serine to facilitate capturing a stable conformation state. Homologs were mainly isolated from human VKOR with some isolated from the pufferfish ''Takifugu rubripes''. Homologs were also tagged at the N and C with a [https:// | + | Structural characterization of VKOR has been difficult due to its in vitro instability. Recently, a series of atomic structures have been determined utilizing anticoagulant stabilization and VKOR-like homologs<ref>DOI 10.1126/science.abc5667</ref>. Crystal structures of VKOR were captured with a bound substrate (KO) or vitamin K antagonist (VKA)<ref>DOI 10.1126/science.abc5667</ref>. VKA substrates utilized were anticoagulants, namely [https://en.wikipedia.org/wiki/Warfarin Warfarin], [https://en.wikipedia.org/wiki/Brodifacoum Brodifacoum], [https://en.wikipedia.org/wiki/Phenindione Phenindione], and [https://en.wikipedia.org/wiki/Chlorophacinone Chlorophacinone]. Second, VKOR-like homologs were utilized to aid in structure classification. Homologs refer to specific cysteine residues that have been mutated to serine to facilitate capturing a stable conformation state. Homologs were mainly isolated from human VKOR with some isolated from the pufferfish ''Takifugu rubripes''. Homologs were also tagged at the N and C with a superfolder [https://en.wikipedia.org/wiki/Green_fluorescent_protein#Applications Green Fluorescent Protein](sfGFP)<ref>DOI 10.1126/science.abc5667</ref>. The [https://proteopedia.org/wiki/index.php/3ed8 sfGFP] provides in vitro stability, a scaffold for crystallization, and facilitates in structure determination. Also, the sfGFP induces states of catalytic activity and potential inhibition for VKOR homologs. For the purpose of this report, all of the structures used have been processed to remove the sfGFP at the south end of VKOR as sfGFP served no purpose in function of the enzyme. This removal allowed for the residue numbering to be reassigned and more closely replicate the human VKOR. |
== Structural Highlights== | == Structural Highlights== | ||
Revision as of 00:45, 19 April 2022
VKOR
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Jin DY, Tie JK, Stafford DW. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry. 2007 Jun 19;46(24):7279-83. doi: 10.1021/bi700527j. Epub 2007 May, 25. PMID:17523679 doi:http://dx.doi.org/10.1021/bi700527j
- ↑ Shen G, Cui W, Cao Q, Gao M, Liu H, Su G, Gross ML, Li W. The catalytic mechanism of vitamin K epoxide reduction in a cellular environment. J Biol Chem. 2021 Jan-Jun;296:100145. doi: 10.1074/jbc.RA120.015401. Epub 2020, Dec 10. PMID:33273012 doi:http://dx.doi.org/10.1074/jbc.RA120.015401
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667
- ↑ Wang Y, Zhang W, Zhang Y, Yang Y, Sun L, Hu S, Chen J, Zhang C, Zheng Y, Zhen Y, Sun K, Fu C, Yang T, Wang J, Sun J, Wu H, Glasgow WC, Hui R. VKORC1 haplotypes are associated with arterial vascular diseases (stroke, coronary heart disease, and aortic dissection). Circulation. 2006 Mar 28;113(12):1615-21. doi: 10.1161/CIRCULATIONAHA.105.580167., Epub 2006 Mar 20. PMID:16549638 doi:http://dx.doi.org/10.1161/CIRCULATIONAHA.105.580167
- ↑ Elshaikh AO, Shah L, Joy Mathew C, Lee R, Jose MT, Cancarevic I. Influence of Vitamin K on Bone Mineral Density and Osteoporosis. Cureus. 2020 Oct 5;12(10):e10816. doi: 10.7759/cureus.10816. PMID:33173624 doi:http://dx.doi.org/10.7759/cureus.10816
- ↑ Patel S, Singh R, Preuss CV, Patel N. Warfarin PMID:29261922
- ↑ Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2020 Nov 5. pii: science.abc5667. doi: 10.1126/science.abc5667. PMID:33154105 doi:http://dx.doi.org/10.1126/science.abc5667