We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1710
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
=== Closed Conformation === | === Closed Conformation === | ||
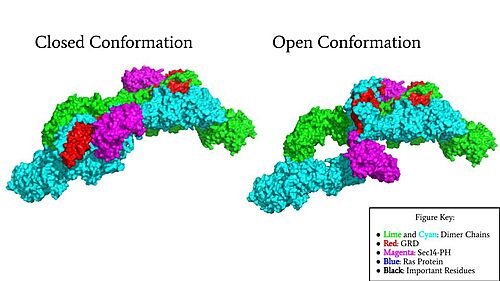
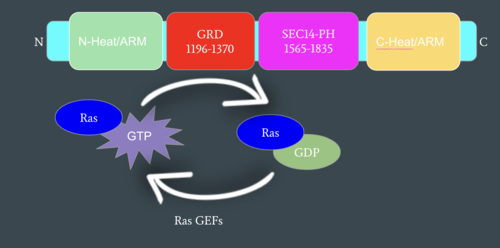
| - | In the <scene name='90/904315/Closed/3'>closed, inactive conformation</scene>, the GRD and Sec14-PH domains are rotated so Ras cannot bind. In this conformation, the GRD and Sec14-PH are inaccessible and inactive. Neurofibromin is held in the inactive state by a <scene name='90/904315/Closed_triade/7'>triad</scene> consisting of residues Cys 1032, His 1558, and His 1576 that form a transition metal-binding site with zinc. The rigid organization of the <scene name='90/904315/Closed_triade/8'>triad in closed conformation</scene> keeps the GRD domain packed tightly on top of the Heat Arms in the Neurofibromin core. This tight compaction sterically occludes Neurofibromin from <scene name='90/904315/Closed_with_ras/2'>associating with Ras.</scene> In its active form, Ras and Neurofibromin will associate via an Arginine Finger(Arg 1276). However, the steric hindrance from the Neurofibromin core in the closed conformation inhibits this association. Therefore, in the closed conformation, neurofibromin cannot catalyze GTP hydrolysis by Ras and Ras continues to signal for cell growth and proliferation.<ref name="Naschberger">PMID:34707296</ref> | + | In the <scene name='90/904315/Closed/3'>closed, inactive conformation</scene>, the GRD and Sec14-PH domains are rotated so Ras cannot bind. In this conformation, the GRD and Sec14-PH are inaccessible and inactive. Neurofibromin is held in the inactive state by a <scene name='90/904315/Closed_triade/7'>triad</scene> consisting of residues Cys 1032, His 1558, and His 1576 that form a transition metal-binding site with zinc. The rigid organization of the <scene name='90/904315/Closed_triade/8'>triad in closed conformation</scene> keeps the GRD domain packed tightly on top of the Heat Arms in the Neurofibromin core. This tight compaction sterically occludes Neurofibromin from <scene name='90/904315/Closed_with_ras/2'>associating with Ras.</scene> In its active form, Ras and Neurofibromin will associate via an <scene name='90/904315/Arg_finger/5'>Arginine Finger</scene>(Arg 1276). However, the <scene name='90/904315/Arg_finger/3'>steric hindrance</scene> from the Neurofibromin core in the closed conformation inhibits this association. Therefore, in the closed conformation, neurofibromin cannot catalyze GTP hydrolysis by Ras and Ras continues to signal for cell growth and proliferation.<ref name="Naschberger">PMID:34707296</ref> |
=== Open Conformation === | === Open Conformation === | ||
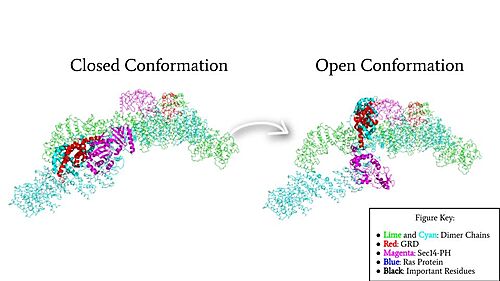
| - | In the <scene name='90/904315/Open_conformation/4'>open, active conformation</scene>, the GRD and Sec14-PH domain on one protomer have [https://youtu.be/I1I4uTVFR00 rotated] and become accessible for binding to Ras. This transition is initiated by the movement of the transition metal-binding site. The Cys 1032, His 1558, and His 1576 Residues become separated and zinc is not able to bond. In the active form, one protomer has its GRD and Sec14-PH domains oriented oppositely from the inactive form (Figure 4). The GRD rotates -130° and the Sec14-PH domain rotates -90° away from the N-HEAT/ARM. Due to this rotation, Cys 1032 is now located too far away, approximately 30 Å, from His 1558 and His 1576 which results in the loss of the metal-binding site and no formation of the <scene name='90/904315/Open_conformation_triade/6'>triad</scene>. The lack of the transition metal-binding site allows the GRD to orient itself to <scene name='90/904315/Openwithras/1'>associate with Ras</scene><ref name="Naschberger">PMID:34707296</ref>. This association positions the Arginine Finger to help stabilize and orient a catalytic Ras residue (Q61) so that the gamma phosphate of GTP can be nucleophilically attacked <ref>PMID:33121128</ref>. When Neurofibromin is in the open, active conformation, | + | In the <scene name='90/904315/Open_conformation/4'>open, active conformation</scene>, the GRD and Sec14-PH domain on one protomer have [https://youtu.be/I1I4uTVFR00 rotated] and become accessible for binding to Ras. This transition is initiated by the movement of the transition metal-binding site. The Cys 1032, His 1558, and His 1576 Residues become separated and zinc is not able to bond. In the active form, one protomer has its GRD and Sec14-PH domains oriented oppositely from the inactive form (Figure 4). The GRD rotates -130° and the Sec14-PH domain rotates -90° away from the N-HEAT/ARM. Due to this rotation, Cys 1032 is now located too far away, approximately 30 Å, from His 1558 and His 1576 which results in the loss of the metal-binding site and no formation of the <scene name='90/904315/Open_conformation_triade/6'>triad</scene>. The lack of the transition metal-binding site allows the GRD to orient itself to <scene name='90/904315/Openwithras/1'>associate with Ras</scene><ref name="Naschberger">PMID:34707296</ref>. This association positions the <scene name='90/904315/Open_conformation_arginine_fin/3'>Arginine Finger</scene> to help stabilize and orient a catalytic Ras residue (Q61) so that the gamma phosphate of GTP can be nucleophilically attacked <ref>PMID:33121128</ref>. When Neurofibromin is in the open, active conformation, <scene name='90/904315/Open_conformation_arginine_fin/4'>Arg1276</scene> is able to bind to Ras because there is no steric hindrance from the Neurofibromin core.<ref name="Naschberger">PMID:34707296</ref> |
[[Image:Domain Rotation.jpg|500 px|left|thumb|Figure 4: Rotation of the GRD and Sec14-PH domains from the closed conformation (7PGR) of neurofibromin to the open conformation (7PGT) of neurofibromin to allow Ras binding. The GRD rotates -130° and the Sec14-PH domain rotates -90°]] | [[Image:Domain Rotation.jpg|500 px|left|thumb|Figure 4: Rotation of the GRD and Sec14-PH domains from the closed conformation (7PGR) of neurofibromin to the open conformation (7PGT) of neurofibromin to allow Ras binding. The GRD rotates -130° and the Sec14-PH domain rotates -90°]] | ||
| Line 37: | Line 37: | ||
*Hannah Luchinski | *Hannah Luchinski | ||
*Sophie Mullinix | *Sophie Mullinix | ||
| - | |||
| - | *<scene name='90/904315/Arg_finger/5'>Arginine Finger Closed zoomed out</scene> | ||
| - | *<scene name='90/904315/Arg_finger/3'>Arginine Finger Closed zoomed in</scene> | ||
| - | *<scene name='90/904315/Open_conformation_arginine_fin/3'>Arginine Finger Open zoomed out</scene> | ||
| - | *<scene name='90/904315/Open_conformation_arginine_fin/4'>Arginine Finger Open zoomed in</scene> | ||
| - | |||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 02:02, 19 April 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Human Neurofibromin - The Tumor Suppressor Gene
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Naschberger A, Baradaran R, Rupp B, Carroni M. The structure of neurofibromin isoform 2 reveals different functional states. Nature. 2021 Nov;599(7884):315-319. doi: 10.1038/s41586-021-04024-x. Epub 2021, Oct 27. PMID:34707296 doi:http://dx.doi.org/10.1038/s41586-021-04024-x
- ↑ 3.0 3.1 Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ 4.0 4.1 4.2 Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015 May;15(5):290-301. doi: 10.1038/nrc3911. Epub 2015 Apr 16. PMID:25877329 doi:http://dx.doi.org/10.1038/nrc3911
- ↑ 5.0 5.1 5.2 Abramowicz A, Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1gene as a cause of disease. Dev Period Med. 2014 Jul-Sep;18(3):297-306. PMID:25182393
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365