BASIL2022GV3R8E
From Proteopedia
| Line 8: | Line 8: | ||
== Introduction == | == Introduction == | ||
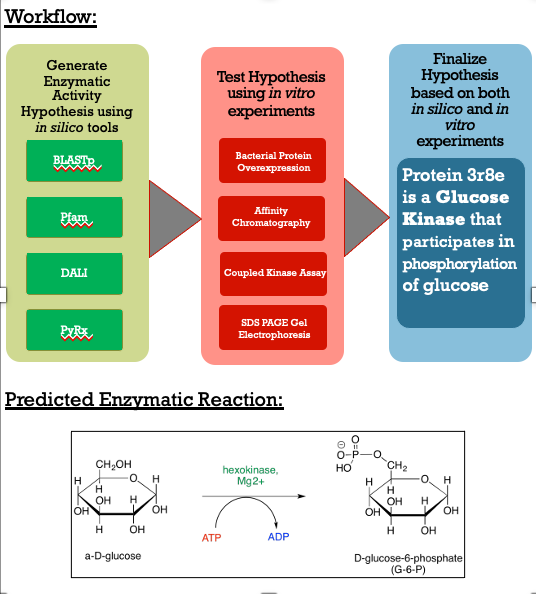
Like many proteins with solved crystal structures, protein 3r8e has an uncharacterized and unconfirmed function. Extracted from and being a part of a bacteria that is found in soil, no one knows what the role of this specific protein plays in relation to the bacteria. Apart of a research project, our group set out to solve the function of this protein to then potentially provide further insight of the protein's relationship to the bacteria. Below is the general workflow of how we got our results. | Like many proteins with solved crystal structures, protein 3r8e has an uncharacterized and unconfirmed function. Extracted from and being a part of a bacteria that is found in soil, no one knows what the role of this specific protein plays in relation to the bacteria. Apart of a research project, our group set out to solve the function of this protein to then potentially provide further insight of the protein's relationship to the bacteria. Below is the general workflow of how we got our results. | ||
| + | |||
[[Image:Workflow.png ]] | [[Image:Workflow.png ]] | ||
== Methods == | == Methods == | ||
Revision as of 22:39, 20 April 2022
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
3. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
4. BASIL. https://basilbiochem.github.io/basil/
5. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
6. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
7. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
8. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky