Methionine synthase
From Proteopedia
| Line 6: | Line 6: | ||
Addlt ref 2<ref>DOI:10.1073/pnas.1133218100</ref> | Addlt ref 2<ref>DOI:10.1073/pnas.1133218100</ref> | ||
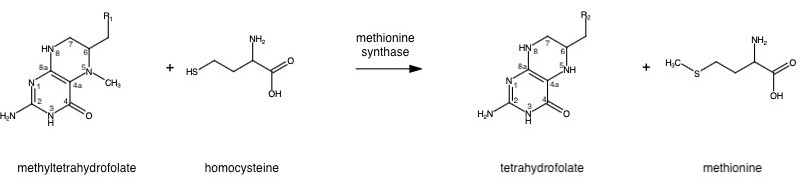
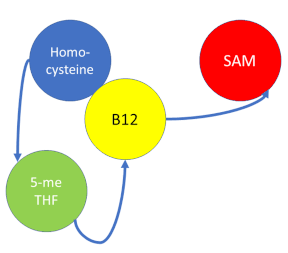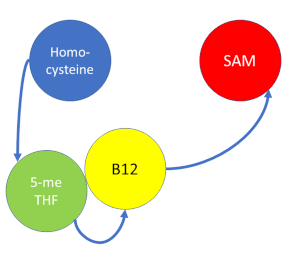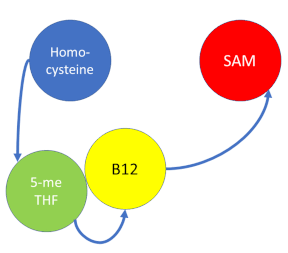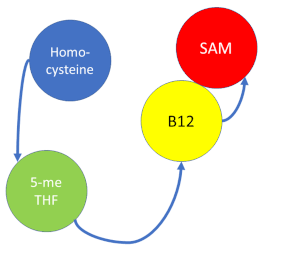
| - | '''Methionine synthase''' (MS; EC: 2.1.1.13) is | + | '''Methionine synthase''' (MS; EC: 2.1.1.13) is the enzyme in [[one-carbon metabolism]] linking the folate and the methionine cycle. MS catalyzes the transfer of a methyl group from methyltetrahydrofolate (MTHF) to homocysteine, resulting in the formation of methionine. Methionine is an essential amino acid required by our bodies for healthy cell and tissue growth. It is essential as it is not naturally derived in our bodies. As it is used as a methyl donor in the form of S-adenosylmethionine, the resulting homocysteine is recyled to form methionine again. |
==Function== | ==Function== | ||
| Line 37: | Line 37: | ||
<StructureSection load='1bmt' size='310' side='right' caption='Homodimer of B12 binding domain of MS. Cobalt in pink.' scene=''> | <StructureSection load='1bmt' size='310' side='right' caption='Homodimer of B12 binding domain of MS. Cobalt in pink.' scene=''> | ||
| - | + | Methionine synthase contains four domains, each with a unique function that bind to Cob(I)alamin as the methyl carrier, MTHF as the methyl donor in the catalytic cycle, homocysteine as the methyl acceptor, and S-adenosylmethionine or SAM, as the methyl donor in the reactivation cycle<ref name="Bandarian et al">DOI: 10.1038/nsb738</ref>. The orientation of the domains changes during the catalytic cycle. | |
| + | [[Image:Methionine synthase domains.gif]] | ||
| + | |||
| + | The full structure of MS has yet to be determined. | ||
Shown here is the <scene name='90/907471/Superposition/7'>theoretical prediction</scene> of the structure by the alphafold algorithm, with experimental structures of the N-terminal 2 domains as well as of the C-terminal 2 domins superposed. | Shown here is the <scene name='90/907471/Superposition/7'>theoretical prediction</scene> of the structure by the alphafold algorithm, with experimental structures of the N-terminal 2 domains as well as of the C-terminal 2 domins superposed. | ||
| Line 73: | Line 76: | ||
The vitamin B12 Cobalamin binding domain has a special characteristic in that, it is most naturally found in a protective conformation to prevent unwanted chemistry from occurring (PDB: 1BMT). This is referred to as a 'capping' mechanism. | The vitamin B12 Cobalamin binding domain has a special characteristic in that, it is most naturally found in a protective conformation to prevent unwanted chemistry from occurring (PDB: 1BMT). This is referred to as a 'capping' mechanism. | ||
| - | <scene name='90/907471/ | + | <scene name='90/907471/Bindingdomain1/1'>scene 1</scene> |
| + | |||
| + | <scene name='90/907471/Bindingdomain2/1'>scene 2</scene> | ||
| - | <scene name='90/907471/Bindingdomain3/2'>scene | + | <scene name='90/907471/Bindingdomain3/2'>scene 3</scene> |
| Line 85: | Line 90: | ||
When B12 is not engaged with one of the other three substrate binding domains, it is protected by a <scene name='90/907471/Cap/1'>cap</scene>. | When B12 is not engaged with one of the other three substrate binding domains, it is protected by a <scene name='90/907471/Cap/1'>cap</scene>. | ||
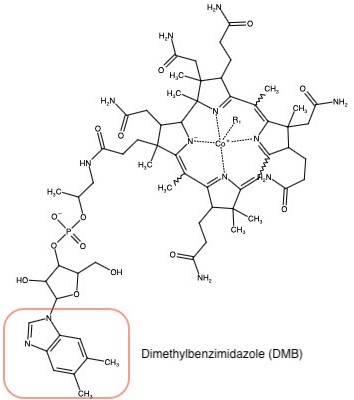
| - | In the Cob(I)alamin binding domain, the imidazole side chain containing His 759 replaces the dimethylbenzimidazole (DMB) ligand. His 759 bonds to Asp 757 and Ser 810 via hydrogen bonds to create a ligand trifecta that increases the efficiency of the methyl transfer during the catalytic cycle. With His on, the cap is off of Cobalamin to allow for it to hold onto the methyl from MTHF. With His off, the cap is on thus no reaction.<ref name="Bandarian et al"/>. | + | In the Cob(I)alamin binding domain, the <scene name='90/907471/Cap/4'>imidazole side chain containing His 759</scene> replaces the dimethylbenzimidazole (DMB) ligand. His 759 bonds to Asp 757 and Ser 810 via hydrogen bonds to create a ligand trifecta that increases the efficiency of the methyl transfer during the catalytic cycle. With His on, the cap is off of Cobalamin to allow for it to hold onto the methyl from MTHF. With His off, the cap is on thus no reaction.<ref name="Bandarian et al"/>. |
Revision as of 16:51, 22 April 2022
This page is being worked on during the Spring 2022 semester.
Addlt ref 1[1]
Addlt ref 2[2]
Methionine synthase (MS; EC: 2.1.1.13) is the enzyme in one-carbon metabolism linking the folate and the methionine cycle. MS catalyzes the transfer of a methyl group from methyltetrahydrofolate (MTHF) to homocysteine, resulting in the formation of methionine. Methionine is an essential amino acid required by our bodies for healthy cell and tissue growth. It is essential as it is not naturally derived in our bodies. As it is used as a methyl donor in the form of S-adenosylmethionine, the resulting homocysteine is recyled to form methionine again.
Contents |
Function
MS is a B12-dependent enzyme responsible for regenerating methionine from homocysteine and uses vitamin B12 Cobalamin as a cofactor. The change from homocysteine to methionine is an SN2 reaction, as seen above, where the methyl group on N-5 from methyltetrahydrofolate (MTHF), is donated. MTHF is a product of methylenetetrahydrofolate reductase MTHFR from the folate cycle. This is a complex reaction as tetrahydrofolate (THF), the product, is a poor leaving group and requires a "super nucleophile", vitamin B12 Cob(I)alamin, to carry out the reaction[3][4]; the methyl carrier.
Oxidation states
Oxidation States of Cobalamin
Cobalamin exists in three different oxidation states during the MS cycle.
Cob(I)alamin: Cobalt in the +1 oxidation state is nicknamed the "super nucleophile" as its high energy is required to carry out the complex SN2 reaction of breaking the bond between THF and the methyl group, in the catalytic cycle.
Co(III)alamin: Cobalt in +3 oxidation state occurs when His 759 replaces the dimethylbenzimidazole (DMB) ligand to allow for the methyl to be accepted by Cob(I)alamin, forming Me-Cob(III)alamin.
Cob(II)alamin: Cob(I)alamin is highlight reactive towards oxygen so occasionally under aerobic conditions, Cob(I)alamin will occasionally undergo oxidation leading to an inactive Cob(II)alamin enzyme in the +2 oxidation state. This is regulated by reductive methylation by using Flavodoxin as an electron donor to reactivate Cob(I)alamin, and subsequently regenerates Me-Cob(III)alamin with a methyl being donated from SAM.
Relevance
MS is an important enzyme responsible for generating methionine, required by our bodies for healthy cell and tissue growth, and protein synthesis. Any MS and/or B12 deficiencies can result in diseases such as abnormal birth defects or anemia[4].
Uses in treating cancer.
Structural highlights
1. Domain organization
| |||||||||||
References
- ↑ Barra L, Fontenelle C, Ermel G, Trautwetter A, Walker GC, Blanco C. Interrelations between glycine betaine catabolism and methionine biosynthesis in Sinorhizobium meliloti strain 102F34. J Bacteriol. 2006 Oct;188(20):7195-204. doi: 10.1128/JB.00208-06. PMID:17015658 doi:http://dx.doi.org/10.1128/JB.00208-06
- ↑ Bandarian V, Ludwig ML, Matthews RG. Factors modulating conformational equilibria in large modular proteins: a case study with cobalamin-dependent methionine synthase. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8156-63. doi:, 10.1073/pnas.1133218100. Epub 2003 Jun 27. PMID:12832615 doi:http://dx.doi.org/10.1073/pnas.1133218100
- ↑ Banerjee R, Ragsdale SW. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev Biochem. 2003;72:209-47. doi: 10.1146/annurev.biochem.72.121801.161828. PMID:14527323 doi:http://dx.doi.org/10.1146/annurev.biochem.72.121801.161828
- ↑ 4.0 4.1 Kung Y, Ando N, Doukov TI, Blasiak LC, Bender G, Seravalli J, Ragsdale SW, Drennan CL. Visualizing molecular juggling within a B(12)-dependent methyltransferase complex. Nature. 2012 Mar 14. doi: 10.1038/nature10916. PMID:22419154 doi:10.1038/nature10916
- ↑ 5.0 5.1 Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nat Struct Biol. 2002 Jan;9(1):53-6. PMID:11731805 doi:10.1038/nsb738
Proteopedia Page Contributors and Editors (what is this?)
Kia Yang, Karsten Theis, Michal Harel, Anna Postnikova, Michael O'Shaughnessy