User:David Gucklhorn/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
]] | ]] | ||
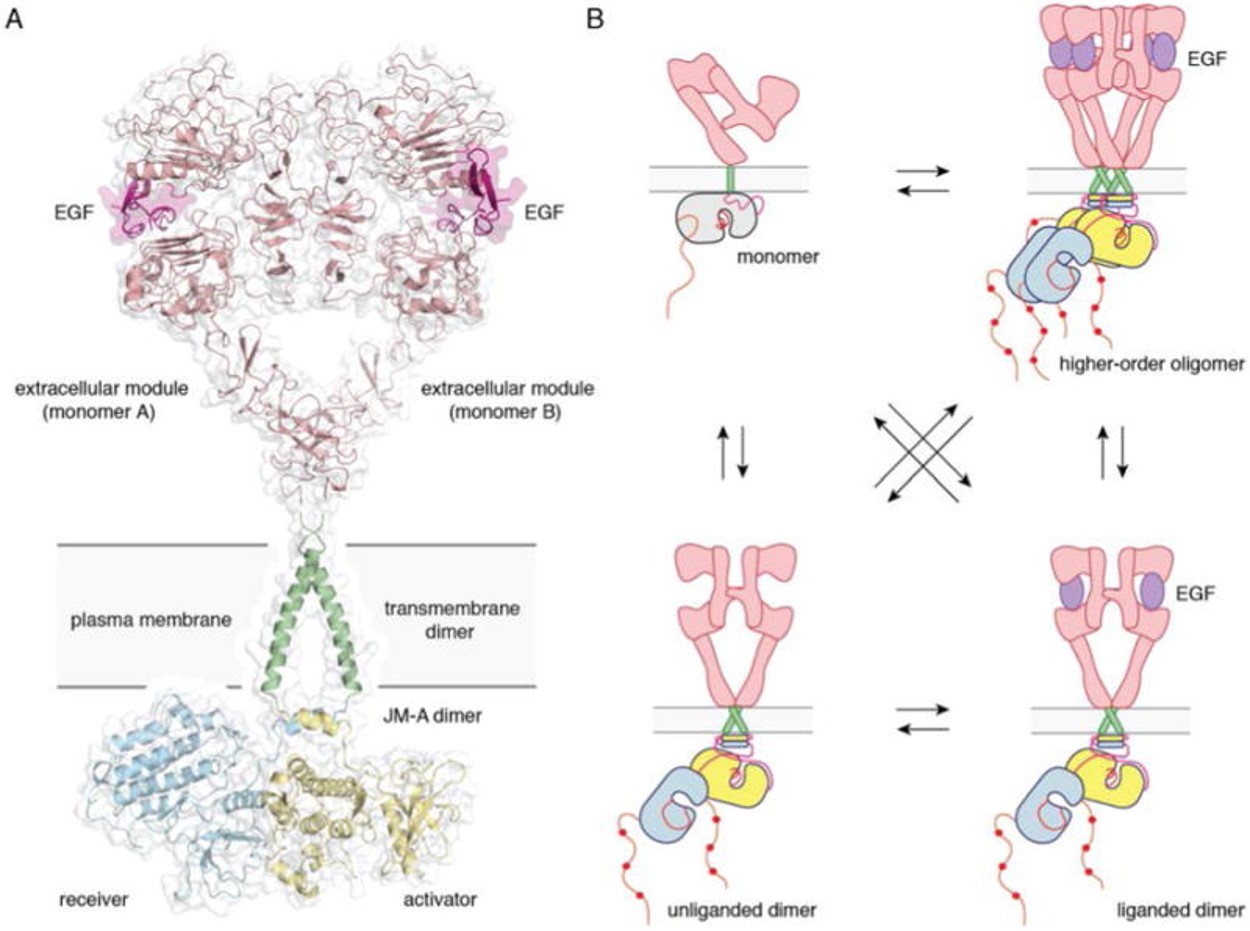
| - | HER2 is an atypical member of the ERBB family, as its ECD adopts an untethered conformation constitutively | + | HER2 is an atypical member of the ERBB family, as its ECD adopts an untethered conformation constitutively <ref>DOI: 10.1016/j.phrs.2013.11.002</ref>. Unlike the other ERBB family members, HER2 does not have a ligand. HER2 preferentially heterodimerizes with ligand bound untethered (open) HER3 or EGFR to initiate cellular signaling, although HER2 homodimers capable of signaling have been reported in HER2 overexpressing cells <ref>DOI: 10.1038/sj.onc.1203967</ref><ref>DOI: 10.1016/j.phrs.2013.11.002</ref><ref>DOI 10.1016/j.ccell.2018.09.010</ref> |
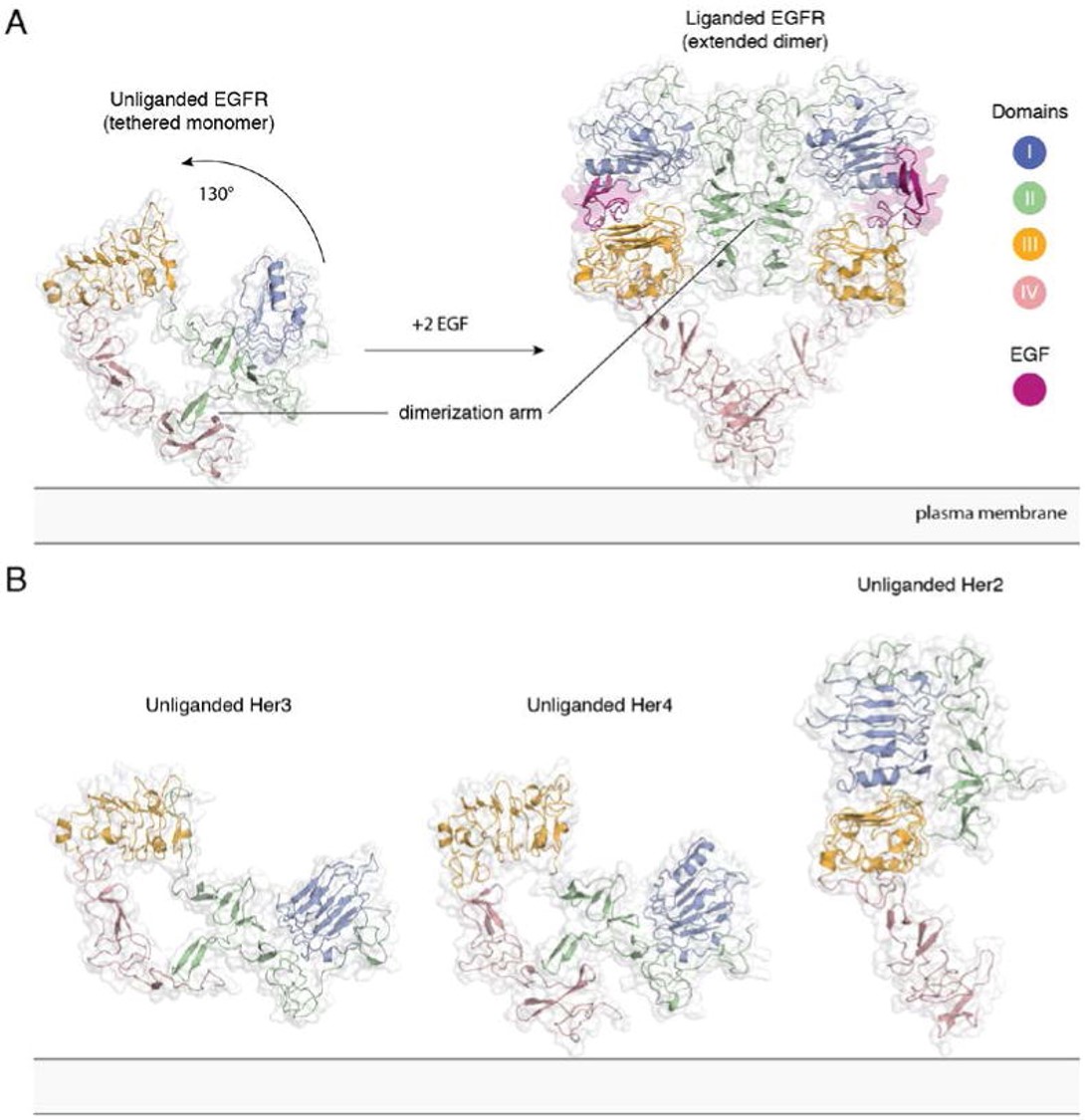
[[Image:Dg_sb_Figure2.jpg|frame|center|Extracellular module structures for the EGFR family members | [[Image:Dg_sb_Figure2.jpg|frame|center|Extracellular module structures for the EGFR family members | ||
A) The conformational change induced by ligand binding. The tethered conformation of EGFR (left, PDB ID 1NQL, EGF bound at low pH was removed for clarity) rearranges to the extended conformation of EGFR (right, PDB ID 3NJP) upon ligand binding. B) Unliganded Her3 (PDB ID 1M6B) and Her4 (PDB ID 2AHX) can adopt a tethered conformation similar to EGFR, while Her2 (PDB ID 1N8H) is in an extended conformation, even in the absence of ligand.<ref>DOI 10.1146/annurev-biochem-060614-034402</ref>]] | A) The conformational change induced by ligand binding. The tethered conformation of EGFR (left, PDB ID 1NQL, EGF bound at low pH was removed for clarity) rearranges to the extended conformation of EGFR (right, PDB ID 3NJP) upon ligand binding. B) Unliganded Her3 (PDB ID 1M6B) and Her4 (PDB ID 2AHX) can adopt a tethered conformation similar to EGFR, while Her2 (PDB ID 1N8H) is in an extended conformation, even in the absence of ligand.<ref>DOI 10.1146/annurev-biochem-060614-034402</ref>]] | ||
| Line 26: | Line 26: | ||
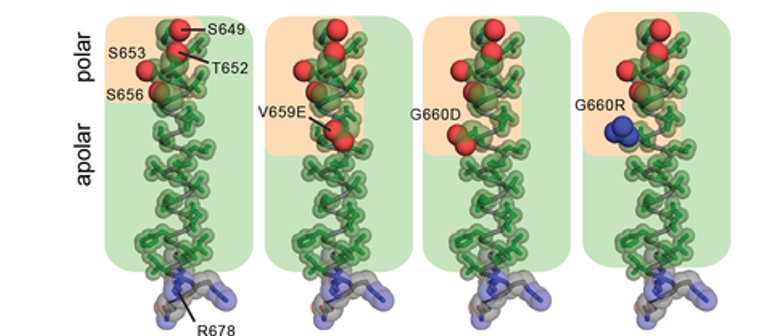
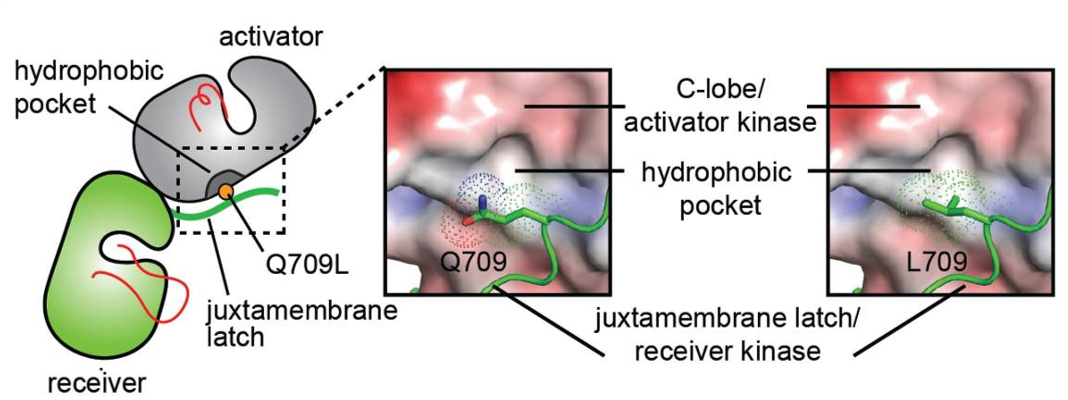
Amino acid composition of the TMD in WT HER2 (PDB ID: 2JWA) and in V659E, G660D, or G660R mutants, highlighting the relative arrangement of side chain atoms of polar (oxygen (red) and nitrogen (blue) atoms shown as spheres) and apolar (carbon atoms (green) shown as sticks) <ref>DOI 10.1016/j.ccell.2018.09.010</ref>]] | Amino acid composition of the TMD in WT HER2 (PDB ID: 2JWA) and in V659E, G660D, or G660R mutants, highlighting the relative arrangement of side chain atoms of polar (oxygen (red) and nitrogen (blue) atoms shown as spheres) and apolar (carbon atoms (green) shown as sticks) <ref>DOI 10.1016/j.ccell.2018.09.010</ref>]] | ||
| - | Proper positioning of the S656-xxx-G660 motif for productive TMD dimer formation is highly dependent on the orientation and geometry of the monomeric TMD helices, defined by basic residues near the interfacial regions between the cytoplasm and head group region of the bilayer | + | Proper positioning of the S656-xxx-G660 motif for productive TMD dimer formation is highly dependent on the orientation and geometry of the monomeric TMD helices, defined by basic residues near the interfacial regions between the cytoplasm and head group region of the bilayer <ref>DOI 10.1007/s00232-010-9323-9</ref><ref>DOI 10.1073/pnas.1215400110</ref><ref>DOI 10.1038/nature10697</ref>. Activating HER2 mutations such as R678Q might have a significant effect on the TMD geometry and dimerization. This was tested by performing all-atom 100 ns MD simulations for wild-type (WT) and the WT/R678Q HER2 TMD dimers in a phospholipid bilayer. The coordinates of the HER2 TM dimer in the putative activated conformation determined by NMR (PDB ID: 2JWA) were used as the starting positions in the simulations. The conformation of the WT HER2 TMD homodimer (WT/WT) remains stable over the course of the simulation. In the WT/R678Q TMD heterodimer, the S656-xxx-G660 motif remained engaged for the duration of the simulation, albeit through different interactions. However, the R678Q containing region of the C-termini separated by several angstroms compared to the WT homodimer (Figure 4). Despite these differences, in both WT/WT and WT/R678Q dimers, the conformations observed in the final state are consistent with a geometry proposed to support an activated, asymmetric configuration of the cytoplasmic kinase domains, and suggests that the enhanced activity of the mutant may be the result of its stabilizing effect on the specific heterodimeric configuration required for signaling. |
| - | A second possibility is that oncogenic mutations are able to stabilize an alternate activated dimeric TMD conformation. It is well known that polar interactions strongly support helix association both in conformations that cooperate with small Sm-xxx-Sm motif dimerization or in entirely unique geometries | + | A second possibility is that oncogenic mutations are able to stabilize an alternate activated dimeric TMD conformation. It is well known that polar interactions strongly support helix association both in conformations that cooperate with small Sm-xxx-Sm motif dimerization or in entirely unique geometries <ref>DOI 10.1126/science.1249783</ref><ref>DOI 10.1038/nature01109</ref><ref>DOI 10.1073/pnas.1003166107</ref>. To see the effect of a polar mutation on HER2 TMD dimers the G660D mutant was simulated. MD simulations demonstrated that the introduction of the protonated aspartate disrupts the native dimeric configuration (Figure 4). In five independent 100 ns simulations, the TMD dimer configuration gradually drifted away from the starting configuration sampled for WT/WT HER2 and without achieving a common final state. On a 100 ns time scale it was not possible, however, to predict with certainty the final geometry of a HER2 TMD dimer in the presence of the G660D/R mutations, but these results suggest that polar mutations at position 660 alter the WT HER2 TMD geometry.<ref>DOI 10.1016/j.ccell.2018.09.010</ref> |
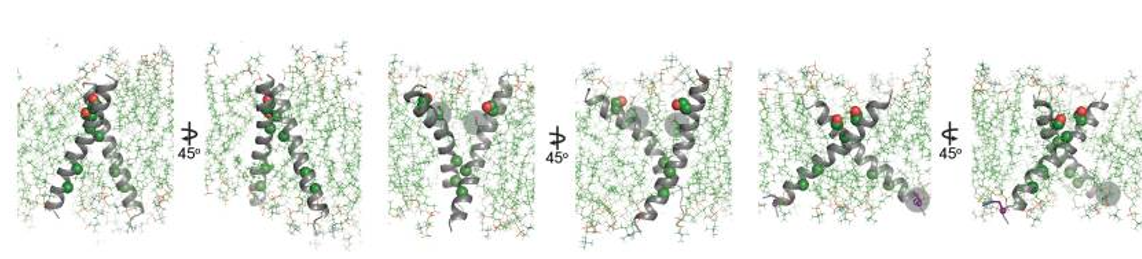
[[Image:Dg_sb_Figure4.png|frame|center|Conformational analysis of HER2 TMD mutants. | [[Image:Dg_sb_Figure4.png|frame|center|Conformational analysis of HER2 TMD mutants. | ||
Overview of the final state obtained at the end of a 100 ns simulation of the WT HER2 TMD dimer (left), a heterodimer between a WT TMD and an activating C-terminal R678Q TMD mutant (WT/R678Q) (middle) and a homodimer of the activating N-terminal G660D TMD mutant (G660D/G660D) (right).<ref>DOI 10.1016/j.ccell.2018.09.010</ref>]] | Overview of the final state obtained at the end of a 100 ns simulation of the WT HER2 TMD dimer (left), a heterodimer between a WT TMD and an activating C-terminal R678Q TMD mutant (WT/R678Q) (middle) and a homodimer of the activating N-terminal G660D TMD mutant (G660D/G660D) (right).<ref>DOI 10.1016/j.ccell.2018.09.010</ref>]] | ||
Revision as of 23:11, 27 April 2022
ErbB2
| |||||||||||
References
- ↑ Graus-Porta D, Beerli RR, Daly JM, Hynes NE. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997 Apr 1;16(7):1647-55. doi: 10.1093/emboj/16.7.1647. PMID:9130710 doi:http://dx.doi.org/10.1093/emboj/16.7.1647
- ↑ Wada T, Qian XL, Greene MI. Intermolecular association of the p185neu protein and EGF receptor modulates EGF receptor function. Cell. 1990 Jun 29;61(7):1339-47. doi: 10.1016/0092-8674(90)90697-d. PMID:1973074 doi:http://dx.doi.org/10.1016/0092-8674(90)90697-d
- ↑ Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006 Jul;7(7):505-16. doi: 10.1038/nrm1962. PMID:16829981 doi:http://dx.doi.org/10.1038/nrm1962
- ↑ Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739-64. doi: 10.1146/annurev-biochem-060614-034402., Epub 2015 Jan 26. PMID:25621509 doi:http://dx.doi.org/10.1146/annurev-biochem-060614-034402
- ↑ Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739-64. doi: 10.1146/annurev-biochem-060614-034402., Epub 2015 Jan 26. PMID:25621509 doi:http://dx.doi.org/10.1146/annurev-biochem-060614-034402
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739-64. doi: 10.1146/annurev-biochem-060614-034402., Epub 2015 Jan 26. PMID:25621509 doi:http://dx.doi.org/10.1146/annurev-biochem-060614-034402
- ↑ Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov , 20. PMID:24269963 doi:http://dx.doi.org/10.1016/j.phrs.2013.11.002
- ↑ Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI. HER2/neu: mechanisms of dimerization/oligomerization. Oncogene. 2000 Dec 11;19(53):6093-101. doi: 10.1038/sj.onc.1203967. PMID:11156522 doi:http://dx.doi.org/10.1038/sj.onc.1203967
- ↑ Roskoski R Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014 Jan;79:34-74. doi: 10.1016/j.phrs.2013.11.002. Epub 2013 Nov , 20. PMID:24269963 doi:http://dx.doi.org/10.1016/j.phrs.2013.11.002
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Kovacs E, Zorn JA, Huang Y, Barros T, Kuriyan J. A structural perspective on the regulation of the epidermal growth factor receptor. Annu Rev Biochem. 2015;84:739-64. doi: 10.1146/annurev-biochem-060614-034402., Epub 2015 Jan 26. PMID:25621509 doi:http://dx.doi.org/10.1146/annurev-biochem-060614-034402
- ↑ Slamon DJ, Clark GM. Amplification of c-erbB-2 and aggressive human breast tumors? Science. 1988 Jun 24;240(4860):1795-8. doi: 10.1126/science.3289120. PMID:3289120 doi:http://dx.doi.org/10.1126/science.3289120
- ↑ Venter DJ, Tuzi NL, Kumar S, Gullick WJ. Overexpression of the c-erbB-2 oncoprotein in human breast carcinomas: immunohistological assessment correlates with gene amplification. Lancet. 1987 Jul 11;2(8550):69-72. doi: 10.1016/s0140-6736(87)92736-x. PMID:2885574 doi:http://dx.doi.org/10.1016/s0140-6736(87)92736-x
- ↑ Fleishman SJ, Schlessinger J, Ben-Tal N. A putative molecular-activation switch in the transmembrane domain of erbB2. Proc Natl Acad Sci U S A. 2002 Dec 10;99(25):15937-40. doi:, 10.1073/pnas.252640799. Epub 2002 Dec 2. PMID:12461170 doi:http://dx.doi.org/10.1073/pnas.252640799
- ↑ Greulich H, Kaplan B, Mertins P, Chen TH, Tanaka KE, Yun CH, Zhang X, Lee SH, Cho J, Ambrogio L, Liao R, Imielinski M, Banerji S, Berger AH, Lawrence MS, Zhang J, Pho NH, Walker SR, Winckler W, Getz G, Frank D, Hahn WC, Eck MJ, Mani DR, Jaffe JD, Carr SA, Wong KK, Meyerson M. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012 Sep 4;109(36):14476-81. doi:, 10.1073/pnas.1203201109. Epub 2012 Aug 20. PMID:22908275 doi:http://dx.doi.org/10.1073/pnas.1203201109
- ↑ Zabransky DJ, Yankaskas CL, Cochran RL, Wong HY, Croessmann S, Chu D, Kavuri SM, Red Brewer M, Rosen DM, Dalton WB, Cimino-Mathews A, Cravero K, Button B, Kyker-Snowman K, Cidado J, Erlanger B, Parsons HA, Manto KM, Bose R, Lauring J, Arteaga CL, Konstantopoulos K, Park BH. HER2 missense mutations have distinct effects on oncogenic signaling and migration. Proc Natl Acad Sci U S A. 2015 Nov 10;112(45):E6205-14. doi:, 10.1073/pnas.1516853112. Epub 2015 Oct 27. PMID:26508629 doi:http://dx.doi.org/10.1073/pnas.1516853112
- ↑ Ross JS, Gay LM, Wang K, Ali SM, Chumsri S, Elvin JA, Bose R, Vergilio JA, Suh J, Yelensky R, Lipson D, Chmielecki J, Waintraub S, Leyland-Jones B, Miller VA, Stephens PJ. Nonamplification ERBB2 genomic alterations in 5605 cases of recurrent and metastatic breast cancer: An emerging opportunity for anti-HER2 targeted therapies. Cancer. 2016 Sep 1;122(17):2654-62. doi: 10.1002/cncr.30102. Epub 2016 Jun 10. PMID:27284958 doi:http://dx.doi.org/10.1002/cncr.30102
- ↑ Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, Monsey J, Goel N, Aronson AB, Li S, Ma CX, Ding L, Mardis ER, Ellis MJ. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013 Feb;3(2):224-37. doi: 10.1158/2159-8290.CD-12-0349. Epub 2012, Dec 7. PMID:23220880 doi:http://dx.doi.org/10.1158/2159-8290.CD-12-0349
- ↑ Yamamoto H, Higasa K, Sakaguchi M, Shien K, Soh J, Ichimura K, Furukawa M, Hashida S, Tsukuda K, Takigawa N, Matsuo K, Kiura K, Miyoshi S, Matsuda F, Toyooka S. Novel germline mutation in the transmembrane domain of HER2 in familial lung adenocarcinomas. J Natl Cancer Inst. 2014 Jan;106(1):djt338. doi: 10.1093/jnci/djt338. Epub 2013, Dec 7. PMID:24317180 doi:http://dx.doi.org/10.1093/jnci/djt338
- ↑ Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, Searleman AC, Shen W, Monsey J, Trusolino L, Jacobs SA, Bertotti A, Bose R. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015 Aug;5(8):832-41. doi: 10.1158/2159-8290.CD-14-1211. PMID:26243863 doi:http://dx.doi.org/10.1158/2159-8290.CD-14-1211
- ↑ Ou SI, Schrock AB, Bocharov EV, Klempner SJ, Haddad CK, Steinecker G, Johnson M, Gitlitz BJ, Chung J, Campregher PV, Ross JS, Stephens PJ, Miller VA, Suh JH, Ali SM, Velcheti V. HER2 Transmembrane Domain (TMD) Mutations (V659/G660) That Stabilize Homo- and Heterodimerization Are Rare Oncogenic Drivers in Lung Adenocarcinoma That Respond to Afatinib. J Thorac Oncol. 2017 Mar;12(3):446-457. doi: 10.1016/j.jtho.2016.11.2224. Epub, 2016 Nov 27. PMID:27903463 doi:http://dx.doi.org/10.1016/j.jtho.2016.11.2224
- ↑ Chang MT, Bhattarai TS, Schram AM, Bielski CM, Donoghue MTA, Jonsson P, Chakravarty D, Phillips S, Kandoth C, Penson A, Gorelick A, Shamu T, Patel S, Harris C, Gao J, Sumer SO, Kundra R, Razavi P, Li BT, Reales DN, Socci ND, Jayakumaran G, Zehir A, Benayed R, Arcila ME, Chandarlapaty S, Ladanyi M, Schultz N, Baselga J, Berger MF, Rosen N, Solit DB, Hyman DM, Taylor BS. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov. 2018 Feb;8(2):174-183. doi: 10.1158/2159-8290.CD-17-0321. Epub, 2017 Dec 15. PMID:29247016 doi:http://dx.doi.org/10.1158/2159-8290.CD-17-0321
- ↑ Petrelli F, Tomasello G, Barni S, Lonati V, Passalacqua R, Ghidini M. Clinical and pathological characterization of HER2 mutations in human breast cancer: a systematic review of the literature. Breast Cancer Res Treat. 2017 Nov;166(2):339-349. doi: 10.1007/s10549-017-4419-x., Epub 2017 Jul 31. PMID:28762010 doi:http://dx.doi.org/10.1007/s10549-017-4419-x
- ↑ Cousin S, Khalifa E, Crombe A, Laizet Y, Lucchesi C, Toulmonde M, Le Moulec S, Auzanneau C, Soubeyran I, Italiano A. Targeting ERBB2 mutations in solid tumors: biological and clinical implications. J Hematol Oncol. 2018 Jun 25;11(1):86. doi: 10.1186/s13045-018-0630-4. PMID:29941010 doi:http://dx.doi.org/10.1186/s13045-018-0630-4
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Hristova K, Wimley WC. A look at arginine in membranes. J Membr Biol. 2011 Jan;239(1-2):49-56. doi: 10.1007/s00232-010-9323-9. Epub 2010 , Nov 25. PMID:21107547 doi:http://dx.doi.org/10.1007/s00232-010-9323-9
- ↑ Gleason NJ, Vostrikov VV, Greathouse DV, Koeppe RE 2nd. Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1692-5. doi:, 10.1073/pnas.1215400110. Epub 2013 Jan 14. PMID:23319623 doi:http://dx.doi.org/10.1073/pnas.1215400110
- ↑ Kim C, Schmidt T, Cho EG, Ye F, Ulmer TS, Ginsberg MH. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. 2011 Dec 18;481(7380):209-13. doi: 10.1038/nature10697. PMID:22178926 doi:10.1038/nature10697
- ↑ Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014 May 16;344(6185):1249783. doi: 10.1126/science.1249783. PMID:24833397 doi:http://dx.doi.org/10.1126/science.1249783
- ↑ Gordeliy VI, Labahn J, Moukhametzianov R, Efremov R, Granzin J, Schlesinger R, Buldt G, Savopol T, Scheidig AJ, Klare JP, Engelhard M. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002 Oct 3;419(6906):484-7. PMID:12368857 doi:10.1038/nature01109
- ↑ Goldberg SD, Clinthorne GD, Goulian M, DeGrado WF. Transmembrane polar interactions are required for signaling in the Escherichia coli sensor kinase PhoQ. Proc Natl Acad Sci U S A. 2010 May 4;107(18):8141-6. doi:, 10.1073/pnas.1003166107. Epub 2010 Apr 19. PMID:20404199 doi:http://dx.doi.org/10.1073/pnas.1003166107
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010
- ↑ Pahuja KB, Nguyen TT, Jaiswal BS, Prabhash K, Thaker TM, Senger K, Chaudhuri S, Kljavin NM, Antony A, Phalke S, Kumar P, Mravic M, Stawiski EW, Vargas D, Durinck S, Gupta R, Khanna-Gupta A, Trabucco SE, Sokol ES, Hartmaier RJ, Singh A, Chougule A, Trivedi V, Dutt A, Patil V, Joshi A, Noronha V, Ziai J, Banavali SD, Ramprasad V, DeGrado WF, Bueno R, Jura N, Seshagiri S. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell. 2018 Nov 12;34(5):792-806.e5. doi: 10.1016/j.ccell.2018.09.010. Epub, 2018 Oct 25. PMID:30449325 doi:http://dx.doi.org/10.1016/j.ccell.2018.09.010