User:Apolena Zounarová/Sandbox 2
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
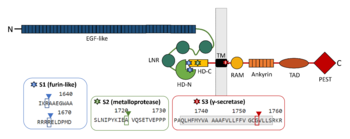
Binding of NOTCH1 ligands such as Delta1 or Jagged1 leads to the dissociation of the heterodimer. This structural change reveals S2 site for a cleavage by metalloprotease TNAα-converting enzyme (TACE), a member of a disintegrin and metalloprotease domain (ADAM) family, which then produces a fragment termed as NOTCH extracellular truncation (NEXT). S2 site is located between A1710 and V1711 in murine NOTCH1, 13 amino acids from the TM domain <ref name="brou">DOI 10.1016/S1097-2765(00)80417-7</ref><ref name="mumm">DOI 10.1016/S1097-2765(00)80416-5</ref>, which corresponds with positions 1720 and 1721 in human NOTCH1 <ref name="uniprot" />. The mechanism of ligand-induced dissociation can be explained by mechanical force caused by simultaneous endocytosis in the ligand cell. Thus, the events in the ligand cell are important for the NOTCH signal transduction as well. Since S2 cleavage is a ligand-regulated step, mutations in heterodimerization domain can mimic ligand-bound stage of the receptor and facilitate NOTCH proteolysis in a similar manner <ref name="nichols">DOI 10.1083/JCB.200609014</ref>. The cleavage by metalloprotease probably brings the receptor in a conformation similar to that of constitutively active receptors <ref name="brou" />. | Binding of NOTCH1 ligands such as Delta1 or Jagged1 leads to the dissociation of the heterodimer. This structural change reveals S2 site for a cleavage by metalloprotease TNAα-converting enzyme (TACE), a member of a disintegrin and metalloprotease domain (ADAM) family, which then produces a fragment termed as NOTCH extracellular truncation (NEXT). S2 site is located between A1710 and V1711 in murine NOTCH1, 13 amino acids from the TM domain <ref name="brou">DOI 10.1016/S1097-2765(00)80417-7</ref><ref name="mumm">DOI 10.1016/S1097-2765(00)80416-5</ref>, which corresponds with positions 1720 and 1721 in human NOTCH1 <ref name="uniprot" />. The mechanism of ligand-induced dissociation can be explained by mechanical force caused by simultaneous endocytosis in the ligand cell. Thus, the events in the ligand cell are important for the NOTCH signal transduction as well. Since S2 cleavage is a ligand-regulated step, mutations in heterodimerization domain can mimic ligand-bound stage of the receptor and facilitate NOTCH proteolysis in a similar manner <ref name="nichols">DOI 10.1083/JCB.200609014</ref>. The cleavage by metalloprotease probably brings the receptor in a conformation similar to that of constitutively active receptors <ref name="brou" />. | ||
Although the cleavage at S2 site is prominent for the activation of the NOTCH pathway <ref name="brou" />, subsequent cleavage by γ-secretase presenilin at S3 site is the one which is responsible for NOTCH intracellular domain (NICD) production <ref name="destrooper">DOI 10.1038/19083</ref>. γ-secretase cleaves murine NOTCH1 between G1743 and V1744 <ref name="schroeter">DOI 10.1038/30756</ref>, which is G1753 and V1754 in human NOTCH1 <ref name="uniprot" />. The same enzymatic activity creates Aβ peptide in Alzheimer‘s disease from β-APP precursor <ref name="destrooper" />. | Although the cleavage at S2 site is prominent for the activation of the NOTCH pathway <ref name="brou" />, subsequent cleavage by γ-secretase presenilin at S3 site is the one which is responsible for NOTCH intracellular domain (NICD) production <ref name="destrooper">DOI 10.1038/19083</ref>. γ-secretase cleaves murine NOTCH1 between G1743 and V1744 <ref name="schroeter">DOI 10.1038/30756</ref>, which is G1753 and V1754 in human NOTCH1 <ref name="uniprot" />. The same enzymatic activity creates Aβ peptide in Alzheimer‘s disease from β-APP precursor <ref name="destrooper" />. | ||
| - | [[Image: | + | [[Image:Figure_Cleavage sites.png|350px|left|thumb| Schematic picture of NOTCH1 domains and cleavage sites with emphasis on regions contributing to the interaction between the two subunits.]] |
===Heterodimerization Domain and Negative Regulatory Region=== | ===Heterodimerization Domain and Negative Regulatory Region=== | ||
Revision as of 16:07, 28 April 2022
NOTCH1 Heterodimerization Domain in T-cell Acute Lymphoblastic Leukaemia
| |||||||||||
References
- ↑ Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587-613. doi:, 10.1146/annurev.pathmechdis.3.121806.154300. PMID:18039126 doi:http://dx.doi.org/10.1146/annurev.pathmechdis.3.121806.154300
- ↑ 2.0 2.1 Gordon WR, Vardar-Ulu D, L'Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009 Aug 24;4(8):e6613. PMID:19701457 doi:10.1371/journal.pone.0006613
- ↑ 3.0 3.1 3.2 . UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021 Jan 8;49(D1):D480-D489. doi: 10.1093/nar/gkaa1100. PMID:33237286 doi:http://dx.doi.org/10.1093/nar/gkaa1100
- ↑ 4.0 4.1 4.2 Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000 Feb;5(2):207-16. doi: 10.1016/s1097-2765(00)80417-7. PMID:10882063 doi:http://dx.doi.org/10.1016/s1097-2765(00)80417-7
- ↑ Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000 Feb;5(2):197-206. doi: 10.1016/s1097-2765(00)80416-5. PMID:10882062 doi:http://dx.doi.org/10.1016/s1097-2765(00)80416-5
- ↑ Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007 Feb 12;176(4):445-58. doi: 10.1083/jcb.200609014. PMID:17296795 doi:http://dx.doi.org/10.1083/jcb.200609014
- ↑ 7.0 7.1 De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999 Apr 8;398(6727):518-22. doi: 10.1038/19083. PMID:10206645 doi:http://dx.doi.org/10.1038/19083
- ↑ Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998 May 28;393(6683):382-6. doi: 10.1038/30756. PMID:9620803 doi:http://dx.doi.org/10.1038/30756
- ↑ Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004 Nov;24(21):9265-73. doi: 10.1128/MCB.24.21.9265-9273.2004. PMID:15485896 doi:http://dx.doi.org/10.1128/MCB.24.21.9265-9273.2004
- ↑ Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996 Feb 20;93(4):1683-8. doi: 10.1073/pnas.93.4.1683. PMID:8643690 doi:http://dx.doi.org/10.1073/pnas.93.4.1683
- ↑ Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008 Mar 22;371(9617):1030-43. doi: 10.1016/S0140-6736(08)60457-2. PMID:18358930 doi:http://dx.doi.org/10.1016/S0140-6736(08)60457-2
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004 Oct 8;306(5694):269-71. doi: 10.1126/science.1102160. PMID:15472075 doi:http://dx.doi.org/10.1126/science.1102160
- ↑ Sundaram M, Greenwald I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics. 1993 Nov;135(3):765-83. PMID:8293978
- ↑ 14.0 14.1 14.2 14.3 14.4 14.5 Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006 Jun;26(12):4642-51. doi: 10.1128/MCB.01655-05. PMID:16738328 doi:http://dx.doi.org/10.1128/MCB.01655-05
- ↑ doi: https://dx.doi.org/10.2210/pdb3I08/pdb
- ↑ Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, Barnes K, Zuurbier L, Meijerink JP, Ferrando AA. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008 Aug 1;112(3):733-40. doi: 10.1182/blood-2007-12-130096. Epub 2008 Apr, 14. PMID:18411416 doi:http://dx.doi.org/10.1182/blood-2007-12-130096
- ↑ Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002 Jun;14(6):637-45. doi: 10.1093/intimm/dxf030. PMID:12039915 doi:http://dx.doi.org/10.1093/intimm/dxf030
- ↑ Schmitt TM, Ciofani M, Petrie HT, Zuniga-Pflucker JC. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J Exp Med. 2004 Aug 16;200(4):469-79. doi: 10.1084/jem.20040394. PMID:15314075 doi:http://dx.doi.org/10.1084/jem.20040394
- ↑ Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell. 1997 Mar 21;88(6):833-43. doi: 10.1016/s0092-8674(00)81929-7. PMID:9118226 doi:http://dx.doi.org/10.1016/s0092-8674(00)81929-7
- ↑ Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006 Jul;20(7):1279-87. doi: 10.1038/sj.leu.2404258. Epub 2006 May 11. PMID:16688224 doi:http://dx.doi.org/10.1038/sj.leu.2404258
- ↑ Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006 Nov 28;103(48):18261-6. doi:, 10.1073/pnas.0606108103. Epub 2006 Nov 17. PMID:17114293 doi:http://dx.doi.org/10.1073/pnas.0606108103
- ↑ Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, Nikov G, Krishnamoorthy V, Majumder PK, Kelliher MA. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009 Jun 11;113(24):6172-81. doi: 10.1182/blood-2008-02-136762. Epub 2009 , Feb 26. PMID:19246562 doi:http://dx.doi.org/10.1182/blood-2008-02-136762
- ↑ Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, Strobl LJ, Honjo T, Clevers H, Radtke F. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 2008 Apr;9(4):377-83. doi: 10.1038/embor.2008.7. Epub 2008 Feb 15. PMID:18274550 doi:http://dx.doi.org/10.1038/embor.2008.7
- ↑ Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, Bhagat G, Agarwal AM, Basso G, Castillo M, Nagase S, Cordon-Cardo C, Parsons R, Zuniga-Pflucker JC, Dominguez M, Ferrando AA. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007 Oct;13(10):1203-10. doi: 10.1038/nm1636. Epub 2007 Sep 16. PMID:17873882 doi:http://dx.doi.org/10.1038/nm1636
- ↑ O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, Draetta G, Sears R, Clurman BE, Look AT. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007 Aug 6;204(8):1813-24. doi: 10.1084/jem.20070876. Epub 2007 Jul, 23. PMID:17646409 doi:http://dx.doi.org/10.1084/jem.20070876
- ↑ Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, Schahin-Reed D, Dow GJ, Shelton A, Stawicki S, Watts RJ, Zhang J, Choy R, Howard P, Kadyk L, Yan M, Zha J, Callahan CA, Hymowitz SG, Siebel CW. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010 Apr 15;464(7291):1052-7. PMID:20393564 doi:10.1038/nature08878
- ↑ Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, Zhao JZ, Baleydier F, L'Heureux S, Wang H, Gordon WR, Thoryk E, Andrawes MB, Tiyanont K, Stegmaier K, Roti G, Ross KN, Franlin LL, Wang H, Wang F, Chastain M, Bett AJ, Audoly LP, Aster JC, Blacklow SC, Huber HE. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010 Feb 8;5(2):e9094. doi: 10.1371/journal.pone.0009094. PMID:20161710 doi:http://dx.doi.org/10.1371/journal.pone.0009094