User:Glauco O. Gavioli Ferreira/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 35: | Line 35: | ||
Maspin does not present the conformational switch already discussed and does not have the consensus motif present in other serpins <ref name="P" />. The intact RCL is necessary for maspin’s activity as a tumor suppressor <ref name="numero11">PMID: 7983035</ref>, but there is no rearrangement of this structure, in other words, there is no S to R conformational change <ref name="numero12">PMID: 7797587</ref><ref name="numero13">PMID: 12384513</ref>. Besides that, the RCL alone has been related to cell matrix adhesion and inhibition of cell invasion <ref name="numero14">PMID: 12799381</ref>. | Maspin does not present the conformational switch already discussed and does not have the consensus motif present in other serpins <ref name="P" />. The intact RCL is necessary for maspin’s activity as a tumor suppressor <ref name="numero11">PMID: 7983035</ref>, but there is no rearrangement of this structure, in other words, there is no S to R conformational change <ref name="numero12">PMID: 7797587</ref><ref name="numero13">PMID: 12384513</ref>. Besides that, the RCL alone has been related to cell matrix adhesion and inhibition of cell invasion <ref name="numero14">PMID: 12799381</ref>. | ||
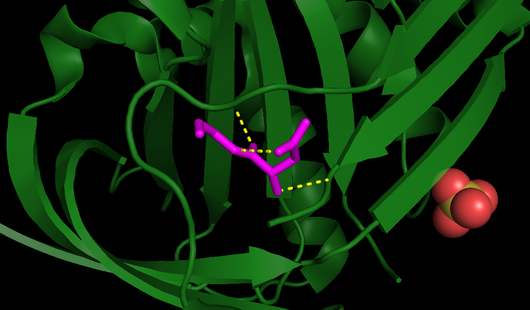
| - | One of the reasons for maspin’s RCL being unable to undergo the conformational switch is its <b>limited flexibility</b>, as it is not flexible like other serpins <ref name="P" />. The RCL of maspin is shorter by four residues and lies closer to the serpin core of the molecule, it is positioned further “back”, in other words closer to the N-terminal, than all of the other known serpin RCL structures. Besides that, the RCL of maspin is also stabilized by bonding interactions with <scene name='91/915218/Betacandrcl/2'>amino acid side chains of the ß-sheet C</scene>, leading to a more rigid structure. Additionally, the breach, where the cleaved RCL is inserted and which is present in other serpins, is not seen on maspin <ref name="numero15">PMID: 15501821</ref>. | + | One of the reasons for maspin’s RCL being unable to undergo the conformational switch is its <b>limited flexibility</b>, as it is not flexible like other serpins <ref name="P" />. The RCL of maspin is shorter by four residues and lies closer to the serpin core of the molecule, it is positioned further “back”, in other words closer to the N-terminal, than all of the other known serpin RCL structures. Besides that, the RCL of maspin is also stabilized by bonding interactions with <scene name='91/915218/Betacandrcl/2'>amino acid side chains of the ß-sheet C</scene> (Fig. 4), leading to a more rigid structure. Additionally, the breach, where the cleaved RCL is inserted and which is present in other serpins, is not seen on maspin <ref name="numero15">PMID: 15501821</ref>. |
| + | |||
| + | [[Image:RCL_lig.png|thumb|right|530x310px|<b>Fig. 4</b> - All RCL bonds, including those responsible for its stability (PDB code [[1wz9]]). RCL is highlighted in <font color='fuchsia'><b>pink</b></font> and all its bonds are displayed as <span style="color:yellow;background-color:black;font-weight:bold;">yellow</span> dashed lines.]] | ||
A curious phenomenon that happens with maspin is the aggregation of <scene name='91/915218/Maspin_dimer/2'>dimers</scene> of tetramers, generating octamers <i>in vitro</i>. There is strong evidence that the hydrophobic residues on the RCL are responsible for the aggregation. The hydrophobic residues Val-336, Ile-341, Leu-342, Pro-337 and adjacent aminoacids present on the RCL are completely exposed to the solvent. Other types of intermolecular interactions, like minor salt links and hydrogen bonds between the s3C and s4C strands of opposing tetramers, also contribute to maintaining the structure, but the main force that results in the octamer are the hydrophobic associations <ref name="numero15" />. Taking into account that the RCL is responsible for functions of cell matrix adhesion and inhibition of cell invasion <ref name="numero14" />, its hydrophobic nature is expected to be functionally important <ref name="numero15" />. | A curious phenomenon that happens with maspin is the aggregation of <scene name='91/915218/Maspin_dimer/2'>dimers</scene> of tetramers, generating octamers <i>in vitro</i>. There is strong evidence that the hydrophobic residues on the RCL are responsible for the aggregation. The hydrophobic residues Val-336, Ile-341, Leu-342, Pro-337 and adjacent aminoacids present on the RCL are completely exposed to the solvent. Other types of intermolecular interactions, like minor salt links and hydrogen bonds between the s3C and s4C strands of opposing tetramers, also contribute to maintaining the structure, but the main force that results in the octamer are the hydrophobic associations <ref name="numero15" />. Taking into account that the RCL is responsible for functions of cell matrix adhesion and inhibition of cell invasion <ref name="numero14" />, its hydrophobic nature is expected to be functionally important <ref name="numero15" />. | ||
Revision as of 07:23, 15 July 2022
SerpinB5 (Maspin)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Khalkhali-Ellis Z. Maspin: the new frontier. Clin Cancer Res. 2006 Dec 15;12(24):7279-83. doi: 10.1158/1078-0432.CCR-06-1589. PMID:17189399 doi:http://dx.doi.org/10.1158/1078-0432.CCR-06-1589
- ↑ 2.0 2.1 Banias L, Jung I, Gurzu S. Subcellular expression of maspin – from normal tissue to tumor cells. World J Meta-Anal 2019; 7(4): 142-155. doi: https://dx.doi.org/10.13105/wjma.v7.i4.142
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Law RH, Irving JA, Buckle AM, Ruzyla K, Buzza M, Bashtannyk-Puhalovich TA, Beddoe TC, Nguyen K, Worrall DM, Bottomley SP, Bird PI, Rossjohn J, Whisstock JC. The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix. J Biol Chem. 2005 Jun 10;280(23):22356-64. Epub 2005 Mar 10. PMID:15760906 doi:http://dx.doi.org/10.1074/jbc.M412043200
- ↑ 4.0 4.1 4.2 4.3 Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. Epub 2006 May 30. PMID:16737556 doi:http://dx.doi.org/10.1186/gb-2006-7-5-216
- ↑ Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000 Oct 19;407(6806):923-6. PMID:11057674 doi:10.1038/35038119
- ↑ Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994 Jan 28;263(5146):526-9. PMID:8290962
- ↑ Goulet B, Kennette W, Ablack A, Postenka CO, Hague MN, Mymryk JS, Tuck AB, Giguere V, Chambers AF, Lewis JD. Nuclear localization of maspin is essential for its inhibition of tumor growth and metastasis. Lab Invest. 2011 Aug;91(8):1181-7. doi: 10.1038/labinvest.2011.66. Epub 2011 Apr , 18. PMID:21502940 doi:http://dx.doi.org/10.1038/labinvest.2011.66
- ↑ 8.0 8.1 Bodenstine TM, Seftor RE, Khalkhali-Ellis Z, Seftor EA, Pemberton PA, Hendrix MJ. Maspin: molecular mechanisms and therapeutic implications. Cancer Metastasis Rev. 2012 Dec;31(3-4):529-51. doi: 10.1007/s10555-012-9361-0. PMID:22752408 doi:http://dx.doi.org/10.1007/s10555-012-9361-0
- ↑ 9.0 9.1 9.2 Yin S, Li X, Meng Y, Finley RL Jr, Sakr W, Yang H, Reddy N, Sheng S. Tumor-suppressive maspin regulates cell response to oxidative stress by direct interaction with glutathione S-transferase. J Biol Chem. 2005 Oct 14;280(41):34985-96. doi: 10.1074/jbc.M503522200. Epub 2005, Jul 26. PMID:16049007 doi:http://dx.doi.org/10.1074/jbc.M503522200
- ↑ Longhi MT, Silva LE, Pereira M, Magalhaes M, Reina J, Vitorino FNL, Gumbiner BM, da Cunha JPC, Cella N. PI3K-AKT, JAK2-STAT3 pathways and cell-cell contact regulate maspin subcellular localization. Cell Commun Signal. 2021 Aug 14;19(1):86. doi: 10.1186/s12964-021-00758-3. PMID:34391444 doi:http://dx.doi.org/10.1186/s12964-021-00758-3
- ↑ Al-Ayyoubi M, Gettins PG, Volz K. Crystal structure of human maspin, a serpin with antitumor properties: reactive center loop of maspin is exposed but constrained. J Biol Chem. 2004 Dec 31;279(53):55540-4. Epub 2004 Oct 22. PMID:15501821 doi:10.1074/jbc.M409957200
- ↑ Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001 Sep 7;276(36):33293-6. doi: 10.1074/jbc.R100016200. Epub 2001, Jul 2. PMID:11435447 doi:http://dx.doi.org/10.1074/jbc.R100016200
- ↑ Sheng S, Pemberton PA, Sager R. Production, purification, and characterization of recombinant maspin proteins. J Biol Chem. 1994 Dec 9;269(49):30988-93. PMID:7983035
- ↑ Pemberton PA, Wong DT, Gibson HL, Kiefer MC, Fitzpatrick PA, Sager R, Barr PJ. The tumor suppressor maspin does not undergo the stressed to relaxed transition or inhibit trypsin-like serine proteases. Evidence that maspin is not a protease inhibitory serpin. J Biol Chem. 1995 Jun 30;270(26):15832-7. PMID:7797587
- ↑ Bass R, Fernandez AM, Ellis V. Maspin inhibits cell migration in the absence of protease inhibitory activity. J Biol Chem. 2002 Dec 6;277(49):46845-8. doi: 10.1074/jbc.C200532200. Epub 2002, Oct 15. PMID:12384513 doi:http://dx.doi.org/10.1074/jbc.C200532200
- ↑ 16.0 16.1 Ngamkitidechakul C, Warejcka DJ, Burke JM, O'Brien WJ, Twining SS. Sufficiency of the reactive site loop of maspin for induction of cell-matrix adhesion and inhibition of cell invasion. Conversion of ovalbumin to a maspin-like molecule. J Biol Chem. 2003 Aug 22;278(34):31796-806. doi: 10.1074/jbc.M302408200. Epub, 2003 Jun 10. PMID:12799381 doi:http://dx.doi.org/10.1074/jbc.M302408200
- ↑ 17.0 17.1 17.2 Al-Ayyoubi M, Gettins PG, Volz K. Crystal structure of human maspin, a serpin with antitumor properties: reactive center loop of maspin is exposed but constrained. J Biol Chem. 2004 Dec 31;279(53):55540-4. Epub 2004 Oct 22. PMID:15501821 doi:10.1074/jbc.M409957200
- ↑ 18.0 18.1 Dzinic SH, Kaplun A, Li X, Bernardo M, Meng Y, Dean I, Krass D, Stemmer P, Shin N, Lonardo F, Sheng S. Identification of an intrinsic determinant critical for maspin subcellular localization and function. PLoS One. 2013 Nov 21;8(11):e74502. doi: 10.1371/journal.pone.0074502., eCollection 2013. PMID:24278104 doi:http://dx.doi.org/10.1371/journal.pone.0074502
- ↑ Li X, Yin S, Meng Y, Sakr W, Sheng S. Endogenous inhibition of histone deacetylase 1 by tumor-suppressive maspin. Cancer Res. 2006 Sep 15;66(18):9323-9. doi: 10.1158/0008-5472.CAN-06-1578. PMID:16982778 doi:http://dx.doi.org/10.1158/0008-5472.CAN-06-1578
- ↑ Bernardo MM, Meng Y, Lockett J, Dyson G, Dombkowski A, Kaplun A, Li X, Yin S, Dzinic S, Olive M, Dean I, Krass D, Moin K, Bonfil RD, Cher M, Sakr W, Sheng S. Maspin reprograms the gene expression profile of prostate carcinoma cells for differentiation. Genes Cancer. 2011 Nov;2(11):1009-22. doi: 10.1177/1947601912440170. PMID:22737267 doi:http://dx.doi.org/10.1177/1947601912440170
- ↑ Lee SJ, Jang H, Park C. Maspin increases Ku70 acetylation and Bax-mediated cell death in cancer cells. Int J Mol Med. 2012 Feb;29(2):225-30. doi: 10.3892/ijmm.2011.833. Epub 2011 Nov, 10. PMID:22076034 doi:http://dx.doi.org/10.3892/ijmm.2011.833
- ↑ Singhal SS, Singh SP, Singhal P, Horne D, Singhal J, Awasthi S. Antioxidant role of glutathione S-transferases: 4-Hydroxynonenal, a key molecule in stress-mediated signaling. Toxicol Appl Pharmacol. 2015 Dec 15;289(3):361-70. doi:, 10.1016/j.taap.2015.10.006. Epub 2015 Oct 23. PMID:26476300 doi:http://dx.doi.org/10.1016/j.taap.2015.10.006
- ↑ Nawata S, Shi HY, Sugino N, Zhang M. Evidence of post-translational modification of the tumor suppressor maspin under oxidative stress. Int J Mol Med. 2011 Feb;27(2):249-54. doi: 10.3892/ijmm.2010.572. Epub 2010 Dec, 2. PMID:21132260 doi:http://dx.doi.org/10.3892/ijmm.2010.572