BASIL2022GV3R8E
From Proteopedia
| Line 24: | Line 24: | ||
For further validation, we conducted an SDS analysis and provided below is the gel image. Results were not as clear as anticipated, and in future studies, we would need to utilize different chromatography methods to yield higher quality protein concentrations and conduct a pre and post induction to visualize the purity of our protein. | For further validation, we conducted an SDS analysis and provided below is the gel image. Results were not as clear as anticipated, and in future studies, we would need to utilize different chromatography methods to yield higher quality protein concentrations and conduct a pre and post induction to visualize the purity of our protein. | ||
| - | + | [[Image:FinalGel.png|500px|]] | |
== Project Implications == | == Project Implications == | ||
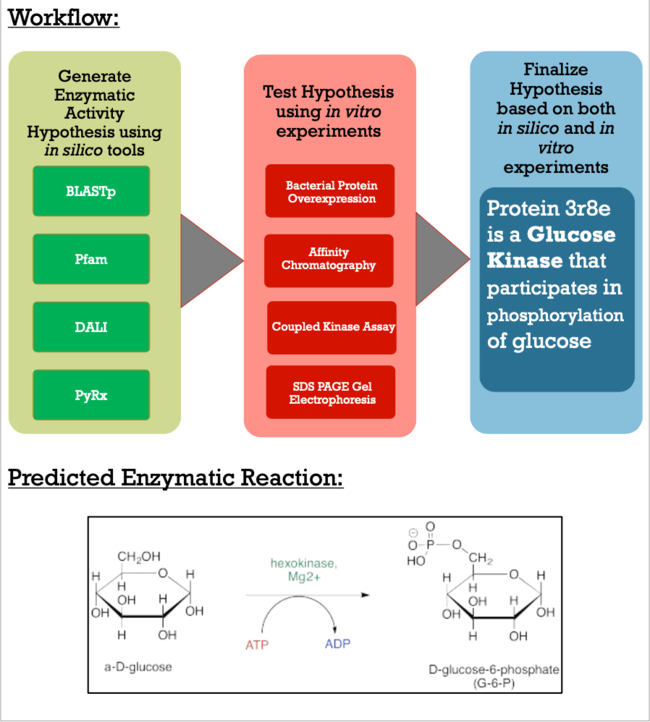
The goal of this project is to explore the methods it takes to characterize a putative kinase. To do this, we became familiar with online alignment, structure, and function tools, paired with a variety of in vitro lab experiments, such as bacterial protein overexpression, affinity chromatography, coupled kinase assays, and SDS PAGE Gel Electrophoresis. Proteins are biomolecules essential to organisms' survival and understanding how they work in result of their function is pivotal for advances in modern medicine, scientific research, and agriculture. | The goal of this project is to explore the methods it takes to characterize a putative kinase. To do this, we became familiar with online alignment, structure, and function tools, paired with a variety of in vitro lab experiments, such as bacterial protein overexpression, affinity chromatography, coupled kinase assays, and SDS PAGE Gel Electrophoresis. Proteins are biomolecules essential to organisms' survival and understanding how they work in result of their function is pivotal for advances in modern medicine, scientific research, and agriculture. | ||
Revision as of 15:57, 21 September 2022
Characterization of the 3r8e Protein, a Novel Glucose Kinase
| |||||||||||
References
1. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
2. BASIL. https://basilbiochem.github.io/basil/
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
5. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky