This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
BASIL2022GV3R8E
From Proteopedia
| Line 4: | Line 4: | ||
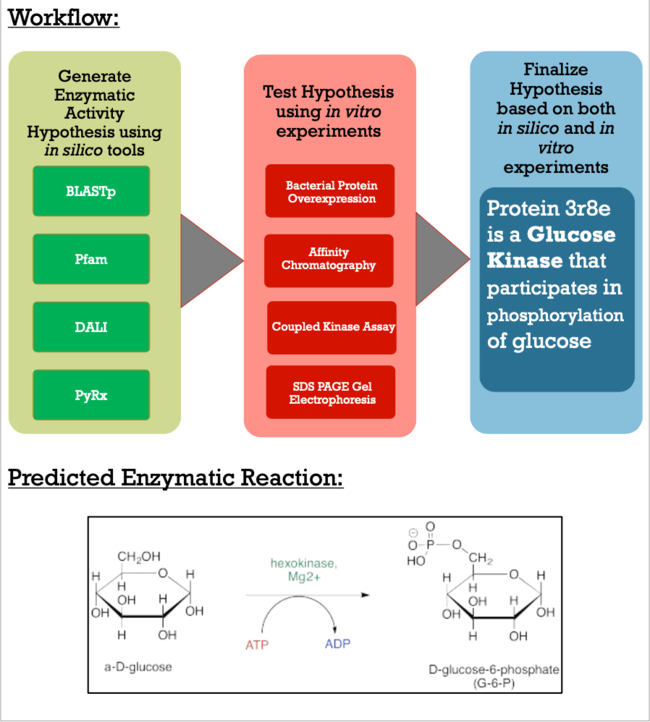
The Protein Data Bank (PDB) contains approximately 188 thousand protein structures, 5000 of which have not been assigned a specific function. As a part of the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) project, we were tasked with analyzing and determining the function of one of these proteins, PDB ID 3r8e. This protein is a putative kinase, which is of interest due to the key roles kinases play in many cellular processes. Utilizing the modules the BASIL consortium provides, a series of in silico and in vitro experiments were conducted. The 3r8e protein was first studied using a variety of in silico tools, including BLASTp, Pfam, and DALI. Based on our in silico results, glucose was determined to be the most likely substrate for 3r8e and was used for further in vitro characterization of the protein. To confirm the in silico function prediction for the 3r8e protein, bacterial protein overexpression, affinity chromatography purification, coupled kinase activity assays, and SDS PAGE analyses were utilized. Multiple sugar substrates for 3r8e were tested, including glucose. The coupled kinase assay results confirmed that 3r8e likely plays a role in glucose phosphorylation, aligning with our in silico conclusions. Previous and subsequent analysis of protein 3r8e validated our initial in silico and in vitro results. Overall, we have strong preliminary evidence that the our protein of interest (POI) is a glucose kinase. | The Protein Data Bank (PDB) contains approximately 188 thousand protein structures, 5000 of which have not been assigned a specific function. As a part of the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) project, we were tasked with analyzing and determining the function of one of these proteins, PDB ID 3r8e. This protein is a putative kinase, which is of interest due to the key roles kinases play in many cellular processes. Utilizing the modules the BASIL consortium provides, a series of in silico and in vitro experiments were conducted. The 3r8e protein was first studied using a variety of in silico tools, including BLASTp, Pfam, and DALI. Based on our in silico results, glucose was determined to be the most likely substrate for 3r8e and was used for further in vitro characterization of the protein. To confirm the in silico function prediction for the 3r8e protein, bacterial protein overexpression, affinity chromatography purification, coupled kinase activity assays, and SDS PAGE analyses were utilized. Multiple sugar substrates for 3r8e were tested, including glucose. The coupled kinase assay results confirmed that 3r8e likely plays a role in glucose phosphorylation, aligning with our in silico conclusions. Previous and subsequent analysis of protein 3r8e validated our initial in silico and in vitro results. Overall, we have strong preliminary evidence that the our protein of interest (POI) is a glucose kinase. | ||
== Introduction == | == Introduction == | ||
| - | As apart of a research project under the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) consortium, our group was tasked with characterizing and identifying the function of this protein to provide further insight of the protein's relationship to the bacteria. Like many proteins with solved crystal structures, protein 3r8e has an uncharacterized and unconfirmed function. Previous research has shown that there is relationship between our POI and bacteria in soil | + | As apart of a research project under the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) consortium, our group was tasked with characterizing and identifying the function of this protein to provide further insight of the protein's relationship to the bacteria. Like many proteins with solved crystal structures, protein 3r8e has an uncharacterized and unconfirmed function. Previous research has shown that there is relationship between our POI and bacteria found in soil. Current research techniques have made the role more apparent and below is the general workflow detailing how we generated our conclusions. |
[[Image:Workflow1.png|650px|]] | [[Image:Workflow1.png|650px|]] | ||
| Line 11: | Line 11: | ||
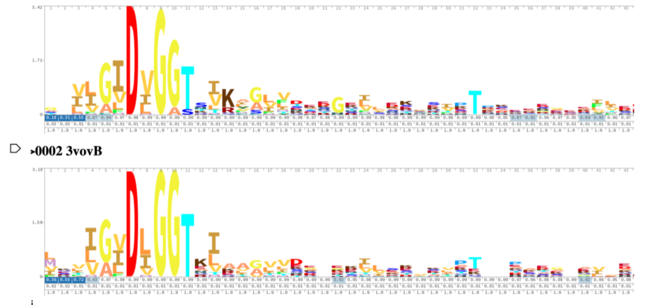
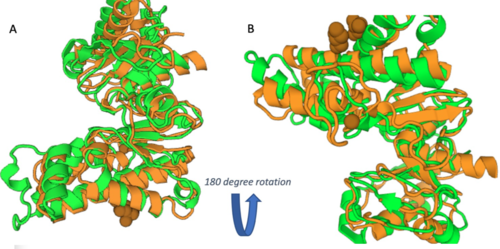
[[Image:3R8EDALIHMM.png|650px|]] [[Image:3R8EDali180.png|500px|]] | [[Image:3R8EDALIHMM.png|650px|]] [[Image:3R8EDali180.png|500px|]] | ||
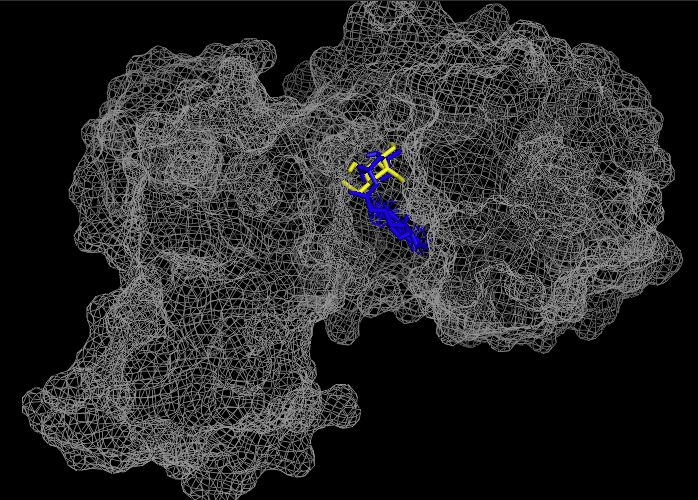
| - | These results, along with supporting evidence from the previous in silico tools, allowed us to continue testing our hypothesis. To begin, we visualized our hypothesized substrate within the active site using mesh view in PyMol. | + | These results, along with supporting evidence from the previous in silico tools, allowed us to continue testing our hypothesis. To begin, we visualized our hypothesized substrate within the active site using mesh view in PyMol. The below PyMol visualization allowed us to see how glucose and ATP interact with our POI. The interactions between glucose (yellow), ATP (blue), and the protein help solidify our initial thoughts. |
[[Image:MeshviewProteinW_atp_glc.png|px|500]] | [[Image:MeshviewProteinW_atp_glc.png|px|500]] | ||
| Line 18: | Line 18: | ||
== Experimental Results/Function == | == Experimental Results/Function == | ||
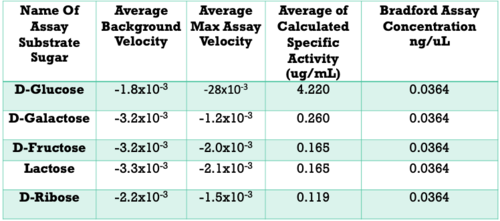
| - | Below are the results of our Uncoupled Kinase Assay. Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. A total of five hexose substrates were tested in vitro, detailed in the table below. Based on these results, we were able to strengthen our initial hypothesis and continue characterization. | + | Once we felt confident enough to finalize our substrate hypothesis, we began testing in vitro. Beginning with bacterial protein overexpression and affinity chromatography, we were able to purify our POI and begin testing with real substrates. Below are the results of our Uncoupled Kinase Assay. Our results from this assay further supports our idea of protein 3r8e assisting in the phosphorylation of glucose. A total of five hexose substrates were tested in vitro, detailed in the table below. Based on these results, we were able to strengthen our initial hypothesis and continue characterization. |
[[Image:KinaseAssayResults.png|500px|]] | [[Image:KinaseAssayResults.png|500px|]] | ||
Revision as of 15:03, 26 September 2022
Characterization of the 3r8e Protein, a Novel Glucose Kinase
| |||||||||||
References
1. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
2. BASIL. https://basilbiochem.github.io/basil/
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
5. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky