This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
BASIL2022GV3R8E
From Proteopedia
| Line 33: | Line 33: | ||
Conclusions: | Conclusions: | ||
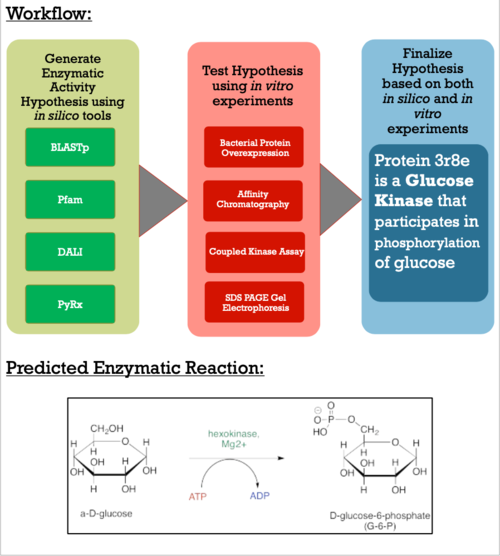
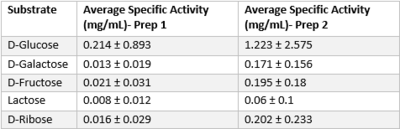
| - | After various experiments and discussion, we have concluded that the novel protein 3R8E is a glucose kinase. By using an in-vitro assay, we obtained results comparing the phosphorylation rates of five sugars in the presence of our putative kinase. From the assay results | + | After various experiments and discussion, we have concluded that the novel protein 3R8E is a glucose kinase. By using an in-vitro assay, we obtained results comparing the phosphorylation rates of five sugars in the presence of our putative kinase. From the assay results, we were able to calculate an average specific activity for all experimental sugars, and by comparing these numbers we can clearly see that glucose is being phosphorylated in the presence of protein 3R8E. To get to this conclusion, we ran triplicates of the experiment for each sugar, and then followed a calculation procedure to get specific activity numbers to quantify how active our protein is with a given substrate. By doing this, we were able to validate and further support our hypothesis, which now can allow others to replicate or continue our research. Glucose provided us with a specific activity value of 0.214 +/- 0.893 and 1.223 +/- 2.575 for prep one and two respectively. |
Future Direction: | Future Direction: | ||
After validating our results, we now look to take our findings to a micropublication website for undergraduate research. By doing this, not only will our work be forward facing and available to the science community, but it also allows for collaboration and further questions to be asked. After completing the micropublication, we look to continue to develop research strategies for putative kinases, as the PDB has thousands of proteins with unsolved functions. We will do this by combining machine learning, data science, and lab work to allow undergraduate students and scientist to effectively research and study putative kinase structures and functions. | After validating our results, we now look to take our findings to a micropublication website for undergraduate research. By doing this, not only will our work be forward facing and available to the science community, but it also allows for collaboration and further questions to be asked. After completing the micropublication, we look to continue to develop research strategies for putative kinases, as the PDB has thousands of proteins with unsolved functions. We will do this by combining machine learning, data science, and lab work to allow undergraduate students and scientist to effectively research and study putative kinase structures and functions. | ||
| - | |||
</StructureSection> | </StructureSection> | ||
Revision as of 19:38, 28 September 2022
Characterization of the 3r8e Protein, a Novel Glucose Kinase
| |||||||||||
References
1. Blastp [Internet]. Bethesda (MD): Natiobal Library of Medicine (US), National Center for Biotechnology Information; 2004- [cited 2022 March]. Available from: (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins)
2. BASIL. https://basilbiochem.github.io/basil/
3. Holm L (2020) Using Dali for protein structure comparison. Methods Mol. Biol. 2112, 29-42.
4. Small- Molecule Library Screening by Docking with PyRx. .Dallakyan S, Olson AJ Methods Mol Biol. 2015;1263:243-50. The full-text is available at https://www.researchgate.net/publications/2739554875. Small-Molecule Library Screening by Docking with PyRx.
5. Pfam: The Protein families database in 2021 J. Mistry, S. Chuguransky, L. Williams, M. Qureshi, G.A. Salazar, E.L.L. Sonnhammer, S.C.E. Tosatto, L. Paladin, S. Raj, L.J. Richardson, R.D. Finn, A. Bateman Nucleic Acids Research (2020) doi: 10.1093/nar/gkaa913
6. The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Dalton Dencklau, Michel Evertsen, Bonnie Hall, Jaime Prilusky