BASIL2023GVQ8DN35
From Proteopedia
| Line 11: | Line 11: | ||
== Function == | == Function == | ||
| - | + | Through computational docking results, the predicted function for the Q8DN35 protein is that of a glucokinase. | |
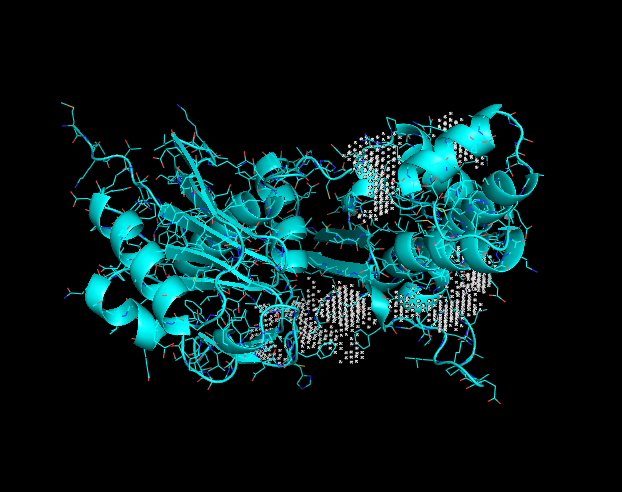
== Binding site == | == Binding site == | ||
Revision as of 15:18, 30 March 2023
Q8DN35 Protein
This is a default text for your page BASIL2023GVQ8DN35. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
|
Contents |
Crystal structure
Function
Through computational docking results, the predicted function for the Q8DN35 protein is that of a glucokinase.
Binding site
This image displays potential binding sites to the Q8DN35 protein.
Protein purification analysis
Through the use of a gravity purification column and Ni-NTA resin, Q8DN35 was purified and a Bradford analysis was conducted to determine the amount of protein in the purified sample. This purification resulted in 0.055 mg/mL of protein being purified.
Kinase activity analysis
To observe Q8DN35's activity, a kinase activity was ran with 2 substrates to observe potential substrate usage. The 2 substrates that were tested are NAG and F-6-P.
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644