This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
SHOC2-MRAS-PP1C, also known as the SMP complex, is a 3-subunit complex essential for cell proliferation and the survival of many cancers and RASopathies. When the subunits comes together, it plays a key role in the activation of the Ras-Raf pathway and signaling cascade. Each subunit of the complex has an individual structure which correlates with its function. SHOC2 has a crescent shape in order to enhance substrate interactions and interactions between the subunits, while PP1C holds the catalytic active site and, the C-terminus of MRAS localizes the complex to the cell membrane. Mutations in one or multiple of these subunits leads to over-activation of the signaling pathway, leading to cancer and developmental disorders called RASopathies. The signaling cascade is kept from over-activating by being held in an auto-inhibited conformation. The SMP complex is responsible for removing this auto-inhibited conformation, allowing for Raf to bind to Ras. Mutations in the subunits can lead to more frequent complex formation, ultimately leading to more cell proliferation. SHOC2-PP1C-MRAS is being studied as a possible treatment target for many types of cancers.
In all images and animations, SHOC2 will be shown as cyan blue, MRAS as lime, and PP1C as violet. Other important components involved in the function of the SMP complex include the 14-3-3 dimer and Raf, which will be shown in salmon and slate-blue, respectively.
Relevance
Cell Proliferation
The Ras-Raf signaling cascade as a whole is fundamental for cell growth and survival. When a membrane bound GTPase is activated by extracellular growth proteins, it binds to a GTP molecule which then activates Raf and the signaling cascade. However, Raf is typically kept in an auto-inhibited form. When MRAS is GTP-bound rather than GDP-bound, it triggers the formation of the SMP complex. The active site of PP1C when in the complex is responsible for removing the residue responsible for auto-inhibition. Extra-cellular growth factors trigger both formation of the SMP complex and Ras-Raf interaction through the binding of GTP to a Ras-protein, however, the SMP complex must remove the auto-inhibition before Ras and Raf can interact. Since SHOC2-PP1C-MRAS plays such a crucial role in the activation of the signaling cascade, many scientists say that cell proliferation is regulated by the SMP complex rather than the Ras-Raf interaction.
Cancer and RASopathies
Mutations in any of the 3 subunits of SHOC2-PP1C-MRAS can lead to cancer or a developmental disability called Rasopathy. Mutations occur at the protein-protein interaction surfaces, leading to more stability of the complex as well as increased interaction energy of SHOC2 with PP1c and/or MRAS. For SHOC2 and PP1C, the mutations lead to amino acid changes on the interaction surfaces, causing a higher affinity for binding. Mutations to MRAS lead to consistent GTP-loading, so the complex is constantly being triggered to form, leading consistent activation of the cell-proliferation pathway even without the presence of the external growth factors. Because the system is no longer regulated, growth of cells when its not needed leads to cancer and/or RASopathies. Furthermore, some mutations in PP1C lead to increased active site enzymatic activity, also leading to increased cell growth.
Structure of Subunits
SHOC2
PP1C
MRAS
Binding of the Subunits
Signaling Cascade
Autoinhibited Confirmation
The first step of the signaling cascade is the dephosphorylation of Raf at Ser259. In the , Raf interacts with a 14-3-3 dimer due to the phosphate group present on Ser259. This interaction with 14-3-3 restrics Raf to the cytoplasm and inhibits Raf from binding with Ras due to steric clash. When GTP binds to MRAS, this triggers the SMP complex to form. Once the complex is formed, PP1C is brought into close proximity of Ras, leading to the dephosphorylation of Ser259. Once dephosphorylated, Raf is in the , allowing for the interaction of Ras and Raf, and the initiation of the signaling cascade.
Switch I and Switch II
SHOC2-PP1C-MRAS is a regulator of a cell proliferation pathway. Mutations in cell proliferation pathways are responsible for 25% of all cancers 1. If this cell proliferation pathway goes unmediated, rapid cell growth and division will occur; the leading cause of cancers is mutations in this pathway. Mechanistic Overview and Signaling Cascade shows the pathway SHOC2-PP1C-MRAS is part of. It is a receptor tyrosine kinase pathway 1. When the receptor binds, a phosphorylation causes a conformational change. This conformation change causes the phosphorylation of other residues. Eventually, this leads to the unbinding of GDP from MRAS and the binding of GTP to MRAS, causing a shift from the open to , The Switch I region is made up of residues 42-48 of the MRAS domain. 1 These residues are crucial for the binding of MRAS, SHOC2, and PP1C. When GDP is bound to the MRAS domain, it is in the . Since the gamma P is not bound to GDP, there are no hydrogen bond interactions with the oxygens of the phosphate group- hence the open conformation. Figure 2 When GTP is bound to MRAS, it is in the closed conformation. The closed conformation allows for the binding of SHOC2 and PP1C. The open conformation of MRAS sterically clashes with the binding site of SHOC2, which is why the complex is not assembled when GDP is bound. The only large-scale conformational change occurs in the MRAS subunit. SHOC2 only undergoes a 6° Since SHOC2 and PP1C do not undergo much conformational change, they are in an equilibrium of binding and unbinding until MRAS-GTP binds to SHOC2, allowing complex assembly.
Ras/Raf
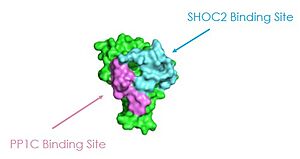
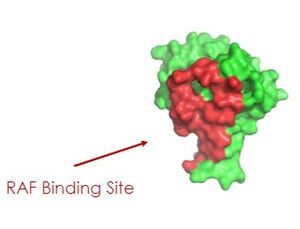
As seen in the figures 1 and 2 below, the surface MRAS uses to recruit SHOC2 and PP1C is the same surface it uses to interact with Raf. As seen in these figures, there is a significant amount of steric overlap with MRAS binding site with PP1C and SHOC2 and Raf. Hence, multiple Ras proteins are required for further activation of the receptor tyrosine kinase pathway. The ability of Ras-GTP to cluster at the membrane is a crucial capability for this protein complex. This anchoring is possible due to the presence of a hydrophobic fatty acid tail on Ras.
Figure 1: MRAS binding sites to SHOC2 and PP1C. This image was created in PyMOL using surface view (PDB code 7DS0).
Figure 2: MRAS binding site to Raf. This image was created in PyMOL using surface view. (PDB code 7DSO). 3
Structure of Active Site
3-Metal Ion Catalysis
The of the SHOC2-PP1C-MRAS complex resides in the PP1C subunit. The role of PP1C is to dephosphorylate SER259 of Raf so that the signaling cascade can start. The active site is unchanged upon the binding of the complex, however, SHOC2 and MRAS aid in the specificity of the enzymatic activity as PP1C is able to dephosphorylate many different targets on its own, with almost 100 PP1C targets found. The full mechanism for the catalytic activity is unknown, however, there are 3 metal ions present (2-Mg2+ and 1-Cl-) to stabilize the waters present in the active site. Additionally, the substrate binds through hydrogen bonds with the main chain and side chain atoms of the catalytic residues. Mutations in the active site lead to increased activity, causing the Ras/Raf signaling cascade to be triggered more frequently.
Hydrophobic Binding Site
PP1C has a adjacent to its active site. The majority of PP1C targets are able to bind through a specific motif that is recognized by the hydrophobic groove. In the Ras/Raf signaling cascade, the region of Raf that is C-terminal to the phosphate group binds to the hydrophobic groove, and the remaining residues bind to the hydrophobic region of SHOC2. This binding to SHOC2 is what allows the SMP complex to be more specific than PP1C on its own.
Future Directions
The knockdown of SHOC2 is being studied as a target for cancer and RASopathy treatment. Although MRAS is the protein that triggers the formation of the complex, SHOC2 is the anchoring location for both MRAS and PP1C. Without SHOC2, the complex would not form and SER259 would not be dephosphorylated. MRAS could be triggered and moved towards the cell membrane, but no complex will form and Raf will remain in the auto-inhibited form. Additionally, there are other RAS proteins that can form an SMP-like complex. If MRAS were to be depleted, other RAS proteins could step in place of MRAS. PP1C is able to dephosphorylated other proteins on it's own, therefore it is not a good target as depletion of PP1C could lead to other issues. Depletion of SHOC2 is the most promising treatment that has been researched. There is also possibility that changing the confirmation of RAS Switch II could lead to decreased cell proliferation.