This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
BASIL2023GVQ8DN35
From Proteopedia
| Line 5: | Line 5: | ||
<Structure load='' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | <Structure load='' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
| - | == | + | == Computational Structure == |
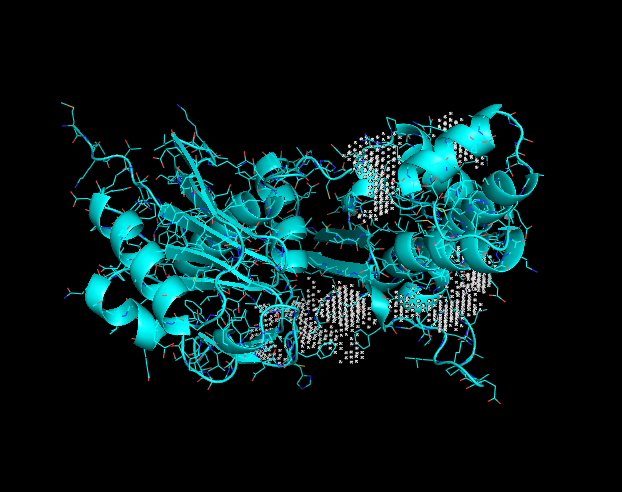
<scene name='95/957644/Q8dn35_structure/2'>Click to display 3D model of Q8DN35</scene> | <scene name='95/957644/Q8dn35_structure/2'>Click to display 3D model of Q8DN35</scene> | ||
Revision as of 17:40, 10 April 2023
Q8DN35 Protein
The predicted function of the Q8DN35 protein was studied during the spring 2023 semester at Grand View University by Lucas Kramer and Anie Rehbein in CHEM 453 - Biochemical Techniques.
|
Contents |
Computational Structure
Function
Through computational docking results, the predicted function for the Q8DN35 protein is that of a glucokinase. The use of tools such as NetGO, BLAST, DALI, InterPRO, PyRX and SPRITE allowed for this predicted function. Q8DN35's sequence is related to that of the ROK Kinase family. NetGO predicted that this protein will have glucokinase activity as it's deepest result. PyRX docking data showed us that NAG (2-acetamido-2-deoxy-beta-D-glucopyranose) and F6P (fructose-6-phosphate) had high binding affinity in the potential binding pocket of Q8DN35.
Binding site
This image displays potential binding sites to the Q8DN35 protein.
Protein purification analysis
Through the use of a gravity purification column and Ni-NTA resin, Q8DN35 was purified and a Bradford analysis was conducted to determine the amount of protein in the purified sample. This purification resulted in 0.055 mg/mL of protein being purified.
Kinase activity analysis
To observe Q8DN35's activity, a kinase activity was ran with 2 substrates to observe potential substrate usage. The 2 substrates that were tested are NAG and F6P.