We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1767
From Proteopedia
(Difference between revisions)
| Line 50: | Line 50: | ||
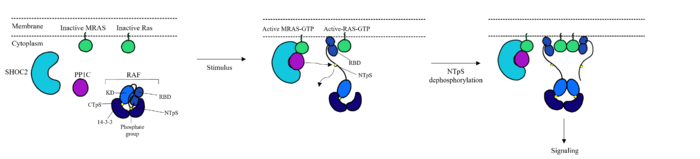
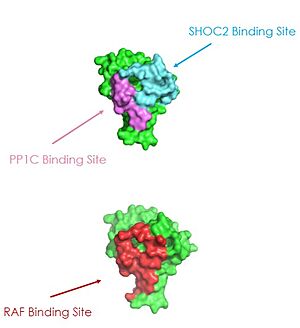
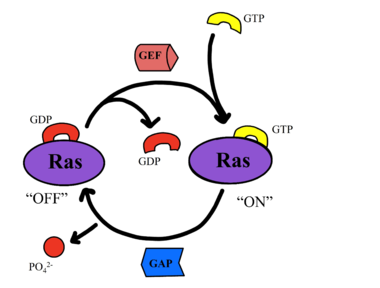
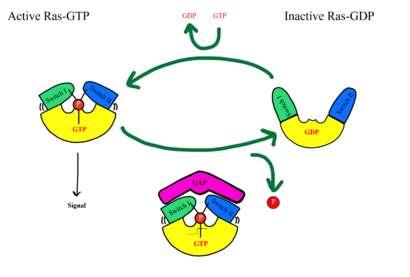
Common mutations in SHOC2 and PP1C lead to amino acid changes on the interaction surfaces, which can result in [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2692211/. higher binding affinity].<ref name="Lavoie">PMID: 35970881</ref>The interface of SHOC2-PP1C is stabilized by the <scene name='95/952695/Q249k_mutation/1'>Q249K</scene> mutation because this creates a salt bridge with E116 of PP1C. This enhances the binding energy by -22.7 kcal/mol. Mutations to MRAS can result in consistent GTP-loading, increasing the formation of the SMP complex in the absence of external growth factors that are necessary for activation of the pathway in a healthy organism. The majority of wild type MRAS in cells are bound to GDP, whereas the MRAS with the Q71L mutation locked MRAS in the GTP bound state.<ref name="Hauseman">PMID:35830882</ref> In MRAS, <scene name='95/952695/Q249k_mutation/2'>Q71L and G23V</scene> both show increased interaction with other effectors such as BRAF, CRAF, and AF6, consistent with gain-of-function mutations that activate MRAS, leading to GTP-loading. | Common mutations in SHOC2 and PP1C lead to amino acid changes on the interaction surfaces, which can result in [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2692211/. higher binding affinity].<ref name="Lavoie">PMID: 35970881</ref>The interface of SHOC2-PP1C is stabilized by the <scene name='95/952695/Q249k_mutation/1'>Q249K</scene> mutation because this creates a salt bridge with E116 of PP1C. This enhances the binding energy by -22.7 kcal/mol. Mutations to MRAS can result in consistent GTP-loading, increasing the formation of the SMP complex in the absence of external growth factors that are necessary for activation of the pathway in a healthy organism. The majority of wild type MRAS in cells are bound to GDP, whereas the MRAS with the Q71L mutation locked MRAS in the GTP bound state.<ref name="Hauseman">PMID:35830882</ref> In MRAS, <scene name='95/952695/Q249k_mutation/2'>Q71L and G23V</scene> both show increased interaction with other effectors such as BRAF, CRAF, and AF6, consistent with gain-of-function mutations that activate MRAS, leading to GTP-loading. | ||
| - | Mutations in PP1C can trigger increased active site activity, increasing the RAF proteins that are active and available to bind to RAS. In patients with [https://medlineplus.gov/genetics/condition/noonan-syndrome/#:~:text=Noonan%20syndrome%20is%20a%20condition,many%20other%20signs%20and%20symptoms. Noonan Syndrome], a disease in the RASopathy family, a point mutation of <scene name='95/952695/Q249k_mutation/2'>T68I</scene> MRAS was identified, | + | Mutations in PP1C can trigger increased active site activity, increasing the RAF proteins that are active and available to bind to RAS. In patients with [https://medlineplus.gov/genetics/condition/noonan-syndrome/#:~:text=Noonan%20syndrome%20is%20a%20condition,many%20other%20signs%20and%20symptoms. Noonan Syndrome], a disease in the RASopathy family, a point mutation of <scene name='95/952695/Q249k_mutation/2'>T68I</scene> MRAS was identified, however the effects this has are unknown.<ref name="Young">PMID: 30348783</ref> Universally, when this MAPK cascade is unregulated, cells are able to proliferate regardless of external signals, leading to [https://www.ncbi.nlm.nih.gov/books/NBK20362/. cancer] and/or RASopathies. |
Current revision
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
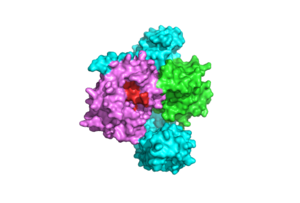
SHOC2-PP1C-MRAS
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Kwon JJ, Hajian B, Bian Y, Young LC, Amor AJ, Fuller JR, Fraley CV, Sykes AM, So J, Pan J, Baker L, Lee SJ, Wheeler DB, Mayhew DL, Persky NS, Yang X, Root DE, Barsotti AM, Stamford AW, Perry CK, Burgin A, McCormick F, Lemke CT, Hahn WC, Aguirre AJ. Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex. Nature. 2022 Jul 13. pii: 10.1038/s41586-022-04928-2. doi:, 10.1038/s41586-022-04928-2. PMID:35831509 doi:http://dx.doi.org/10.1038/s41586-022-04928-2
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Hauseman ZJ, Fodor M, Dhembi A, Viscomi J, Egli D, Bleu M, Katz S, Park E, Jang DM, Porter KA, Meili F, Guo H, Kerr G, Molle S, Velez-Vega C, Beyer KS, Galli GG, Maira SM, Stams T, Clark K, Eck MJ, Tordella L, Thoma CR, King DA. Structure of the MRAS-SHOC2-PP1C phosphatase complex. Nature. 2022 Jul 13. pii: 10.1038/s41586-022-05086-1. doi:, 10.1038/s41586-022-05086-1. PMID:35830882 doi:http://dx.doi.org/10.1038/s41586-022-05086-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J. Structural basis for SHOC2 modulation of RAS signalling. Nature. 2022 Jun 29. pii: 10.1038/s41586-022-04838-3. doi:, 10.1038/s41586-022-04838-3. PMID:35768504 doi:http://dx.doi.org/10.1038/s41586-022-04838-3
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 Lavoie H, Therrien M. Structural keys unlock RAS-MAPK cellular signalling pathway. Nature. 2022 Sep;609(7926):248-249. PMID:35970881 doi:10.1038/d41586-022-02189-7
- ↑ 5.0 5.1 Young LC, Hartig N, Boned Del Río I, Sari S, Ringham-Terry B, Wainwright JR, Jones GG, McCormick F, Rodriguez-Viciana P. SHOC2-MRAS-PP1 complex positively regulates RAF activity and contributes to Noonan syndrome pathogenesis. Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):E10576-E10585. PMID:30348783 doi:10.1073/pnas.1720352115
- ↑ Kelker MS, Page R, Peti W. Crystal structures of protein phosphatase-1 bound to nodularin-R and tautomycin: a novel scaffold for structure-based drug design of serine/threonine phosphatase inhibitors. J Mol Biol. 2009 Jan 9;385(1):11-21. Epub 2008 Nov 1. PMID:18992256 doi:10.1016/j.jmb.2008.10.053
Student Contributors
- Sloan August
- Rosa Trippel
- Kayla Wilhoite