User:Matheus Andrade Bettiol/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
RhoA (Ras homology gene family member A) is a protein of the small GTPase family. It can be in two conformations, <scene name='97/973102/Rhoa_gtp/1'>linked to GTP</scene> and therefore active, or <scene name='97/973102/Rhoa_gdp/4'>linked to GDP</scene> and consequently inactive. Three factors regulate these two states: | RhoA (Ras homology gene family member A) is a protein of the small GTPase family. It can be in two conformations, <scene name='97/973102/Rhoa_gtp/1'>linked to GTP</scene> and therefore active, or <scene name='97/973102/Rhoa_gdp/4'>linked to GDP</scene> and consequently inactive. Three factors regulate these two states: | ||
| - | 1. GEF (Guanine nucleotide exchange factors): promotes the exchange of GDP for GTP, activating RhoA | + | '''1.''' GEF (Guanine nucleotide exchange factors): promotes the exchange of GDP for GTP, activating RhoA |
| - | 2. GAP (GTPase activating proteins): accelerates the hydrolysis of GTP, inhibiting RhoA | + | '''2.''' GAP (GTPase activating proteins): accelerates the hydrolysis of GTP, inhibiting RhoA |
| - | 3. GDI ([[Guanine nucleotide dissociation inhibitor]]): translocates the membrane GTPase, sequestering it to the cytosol, also inhibiting RhoA | + | '''3.''' GDI ([[Guanine nucleotide dissociation inhibitor]]): translocates the membrane GTPase, sequestering it to the cytosol, also inhibiting RhoA |
| Line 48: | Line 48: | ||
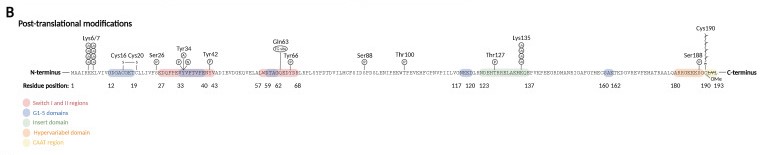
== Post-Translational Modifications == | == Post-Translational Modifications == | ||
| - | Prenylation: the activation of [[Rho GTPase]] require membrane binding, which is necessary for the interaction with membranous GEFs. The membrane association requires C-terminal prenylation, which involves the addition of a geranylgeranyl (20-carbon chain) to Cys190 in the CAAX motif. | + | '''Prenylation:''' the activation of [[Rho GTPase]] require membrane binding, which is necessary for the interaction with membranous GEFs. The membrane association requires C-terminal prenylation, which involves the addition of a geranylgeranyl (20-carbon chain) to Cys190 in the CAAX motif. |
| - | Phosphorylation: can alter the subcellular localization of RhoA when occurs close to C-terminal lipid modifications. On the other hand, phosphorylation of the G-domain affects GTP/GDP cycling and the interaction with effector proteins. | + | '''Phosphorylation:''' can alter the subcellular localization of RhoA when occurs close to C-terminal lipid modifications. On the other hand, phosphorylation of the G-domain affects GTP/GDP cycling and the interaction with effector proteins. |
| - | Oxidation: RhoA can be oxidized on Cys16 and Cys20 (G1 domain), generating a disulfide bond that prevents guanine binding and GEF association, inactivating RhoA. However, if Tyr42 is phosphorylated, serving as a binding site for GEF, oxidation on Cys16/20 reduces the affinity of RhoA for GDI and increases the association with GEF, leading to RhoA activation. | + | '''Oxidation:''' RhoA can be oxidized on Cys16 and Cys20 (G1 domain), generating a disulfide bond that prevents guanine binding and GEF association, inactivating RhoA. However, if Tyr42 is phosphorylated, serving as a binding site for GEF, oxidation on Cys16/20 reduces the affinity of RhoA for GDI and increases the association with GEF, leading to RhoA activation. |
| - | Nitration: nitration on RhoA's Tyr34 (switch I region) introduces a negative charge that modifies the protein structure and leads to a faster GDP release and GTP reload, increasing RhoA activity. | + | '''Nitration:''' nitration on RhoA's Tyr34 (switch I region) introduces a negative charge that modifies the protein structure and leads to a faster GDP release and GTP reload, increasing RhoA activity. |
| - | Adenylation: adenylation on Tyr34 (switch I region) leads to RhoA inhibition. | + | '''Adenylation:''' adenylation on Tyr34 (switch I region) leads to RhoA inhibition. |
Ubiquitination: target the protein for degradation by the proteasome. RhoA is ubiquitylated by E3 [[ubiquitin protein ligase]] complexes, that ubiquitinate either active RhoA on Lys6 and Lys7, inactive RhoA, or both states on Lys135. | Ubiquitination: target the protein for degradation by the proteasome. RhoA is ubiquitylated by E3 [[ubiquitin protein ligase]] complexes, that ubiquitinate either active RhoA on Lys6 and Lys7, inactive RhoA, or both states on Lys135. | ||
[[Image:Pos-translational_modifications_RhoA.jpg]] | [[Image:Pos-translational_modifications_RhoA.jpg]] | ||
| - | Image from | + | Image from Schmidt SI, Blaabjerg M, Freude K, Meyer M. RhoA Signaling in Neurodegenerative Diseases. Cells. 2022 May 1;11(9):1520. |
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">Secondary Structure</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">Secondary Structure</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 21:30, 25 June 2023
==rhoA==
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644