User:Matheus Andrade Bettiol/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='1A2B' size='340' side='right' caption='3D rhoA GTP structure' scene=''> | <StructureSection load='1A2B' size='340' side='right' caption='3D rhoA GTP structure' scene=''> | ||
| - | RhoA (Ras homology gene family member A) is a protein of the small GTPase family. It can be in two conformations, <scene name='97/973102/Rhoa_gtp/1'>linked to GTP</scene> and therefore active, or <scene name='97/973102/Rhoa_gdp/4'>linked to GDP</scene> and consequently inactive. Three factors regulate these two states: | + | RhoA (Ras homology gene family member A) is a protein of the small GTPase family. It can be in two conformations, <scene name='97/973102/Rhoa_gtp/1'>linked to GTP</scene> and therefore active, or <scene name='97/973102/Rhoa_gdp/4'>linked to GDP</scene> and consequently inactive. Three factors regulate these two states <ref>PMID:16212495</ref>: |
'''1.''' GEF (Guanine nucleotide exchange factors): promotes the exchange of GDP for GTP, activating RhoA | '''1.''' GEF (Guanine nucleotide exchange factors): promotes the exchange of GDP for GTP, activating RhoA | ||
| Line 19: | Line 19: | ||
== Disease == | == Disease == | ||
| - | Mutations in RHOA have been linked to a predisposition to autoimmune diseases and cancer progression. Additionally, RhoA signaling is possibly involved in the pathogenesis of neurodegenerative diseases, including Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS). This may be related to the implication of Rho GTPases in brain development since these neurodegenerative diseases present an abnormal accumulation of misfolded peptides. One of these proteins could be RhoA. | + | Mutations in RHOA have been linked to a predisposition to autoimmune diseases and cancer progression <ref>PMID:27138333</ref>. Additionally, RhoA signaling is possibly involved in the pathogenesis of neurodegenerative diseases, including Parkinson’s disease (PD), Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS) <ref>PMID:35563826</ref>. This may be related to the implication of Rho GTPases in brain development since these neurodegenerative diseases present an abnormal accumulation of misfolded peptides. One of these proteins could be RhoA. |
In addition, different bacteria use a pathogenic strategy of inactivating RhoA through their toxins, which make post-translational modifications in the switch I region. The bacteria and their respective toxins are: | In addition, different bacteria use a pathogenic strategy of inactivating RhoA through their toxins, which make post-translational modifications in the switch I region. The bacteria and their respective toxins are: | ||
- Vibrio parahaemolyticus: VopS (adenylation of Thr 37) | - Vibrio parahaemolyticus: VopS (adenylation of Thr 37) | ||
| Line 29: | Line 29: | ||
- Clostridium difficile: TcdB/A (glycosylation of Thr 37) | - Clostridium difficile: TcdB/A (glycosylation of Thr 37) | ||
| - | - Burkholderia cenocepacia: deamidation of Asn 41 | + | - Burkholderia cenocepacia: deamidation of Asn 41 <ref>PMID:24919149</ref> |
| Line 57: | Line 57: | ||
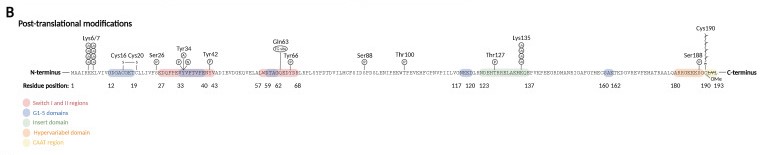
'''Adenylation:''' adenylation on Tyr34 (switch I region) leads to RhoA inhibition. | '''Adenylation:''' adenylation on Tyr34 (switch I region) leads to RhoA inhibition. | ||
| - | Ubiquitination: target the protein for degradation by the proteasome. RhoA is ubiquitylated by E3 [[ubiquitin protein ligase]] complexes, that ubiquitinate either active RhoA on Lys6 and Lys7, inactive RhoA, or both states on Lys135. | + | Ubiquitination: target the protein for degradation by the proteasome. RhoA is ubiquitylated by E3 [[ubiquitin protein ligase]] complexes, that ubiquitinate either active RhoA on Lys6 and Lys7, inactive RhoA, or both states on Lys135.<ref>PMID:35563826</ref> |
[[Image:Pos-translational_modifications_RhoA.jpg]] | [[Image:Pos-translational_modifications_RhoA.jpg]] | ||
Revision as of 21:37, 25 June 2023
==rhoA==
| |||||||||||
References
- ↑ Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247-69. PMID:16212495 doi:10.1146/annurev.cellbio.21.020604.150721
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Bros M, Haas K, Moll L, Grabbe S. RhoA as a Key Regulator of Innate and Adaptive Immunity. Cells. 2019 Jul 17;8(7):733. PMID:31319592 doi:10.3390/cells8070733
- ↑ Hetmanski JH, Zindy E, Schwartz JM, Caswell PT. A MAPK-Driven Feedback Loop Suppresses Rac Activity to Promote RhoA-Driven Cancer Cell Invasion. PLoS Comput Biol. 2016 May 3;12(5):e1004909. PMID:27138333 doi:10.1371/journal.pcbi.1004909
- ↑ Schmidt SI, Blaabjerg M, Freude K, Meyer M. RhoA Signaling in Neurodegenerative Diseases. Cells. 2022 May 1;11(9):1520. PMID:35563826 doi:10.3390/cells11091520
- ↑ Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong YN, Peng X, Xi JJ, Chen S, Wang F, Shao F. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014 Sep 11;513(7517):237-41. doi: 10.1038/nature13449. Epub 2014 Jun 11. PMID:24919149 doi:http://dx.doi.org/10.1038/nature13449
- ↑ Schmidt SI, Blaabjerg M, Freude K, Meyer M. RhoA Signaling in Neurodegenerative Diseases. Cells. 2022 May 1;11(9):1520. PMID:35563826 doi:10.3390/cells11091520