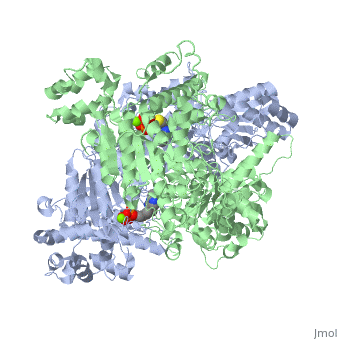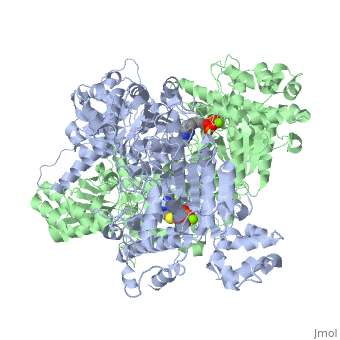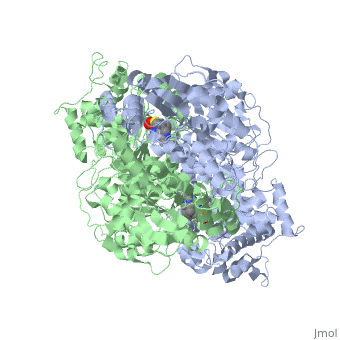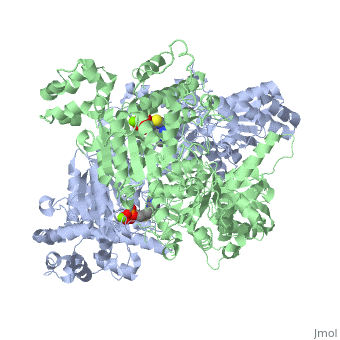
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Pyruvate dehydrogenase
From Proteopedia
(Difference between revisions)
m (Typo: residue 262 coordinating Mg is glutamine, not glutamate) |
|||
| Line 21: | Line 21: | ||
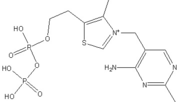
The enzyme requires <scene name='Kenny_Coggins_Sandbox_1/Thdp/5'>TPP and Mg2+</scene> as cofactors for catalysis. [[Image:Thiamine pyrophosphate.png|thumb|left|alt=Thiamine pyrophosphate.|Thiamine pyrophosphate (TPP) E1 cofactor.]] | The enzyme requires <scene name='Kenny_Coggins_Sandbox_1/Thdp/5'>TPP and Mg2+</scene> as cofactors for catalysis. [[Image:Thiamine pyrophosphate.png|thumb|left|alt=Thiamine pyrophosphate.|Thiamine pyrophosphate (TPP) E1 cofactor.]] | ||
{{Clear}} | {{Clear}} | ||
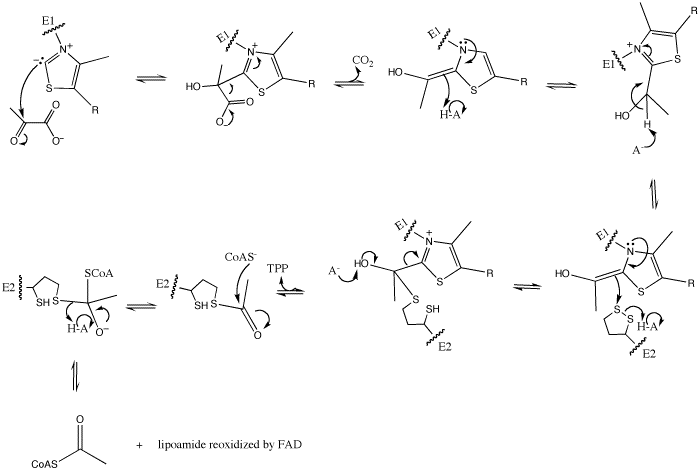
| - | In this reaction, the ylide form of TPP attacks the electrophilic carbonyl group of pyruvate. This reflects the ability of TPP’s thiazolium ring, which primarily interacts with <scene name='Kenny_Coggins_Sandbox_1/Ile569_asp521/5'>Ile569 and Asp521</scene>, to add to carbonyl groups. Decarboxylation of the resulting alkoxide yields an enol complex. This enol resonates to form the ylide form of hydroxyethyl-TPP<ref name="book">Voet, D., Voet, J.G., and Pratt, C.W. (2008). Fundamentals of biochemistry. Hoboken, NJ: John Wiley and Sons, Inc.</ref>. During this reaction, the catalytic Mg2+ ion coordinates octahedrally with three protein ligands: <scene name='Kenny_Coggins_Sandbox_1/Asp230_and_asn260/2'>Asp230 and Asn 260</scene>, which bind TPP, and <scene name='Kenny_Coggins_Sandbox_1/Glu262/2'> | + | In this reaction, the ylide form of TPP attacks the electrophilic carbonyl group of pyruvate. This reflects the ability of TPP’s thiazolium ring, which primarily interacts with <scene name='Kenny_Coggins_Sandbox_1/Ile569_asp521/5'>Ile569 and Asp521</scene>, to add to carbonyl groups. Decarboxylation of the resulting alkoxide yields an enol complex. This enol resonates to form the ylide form of hydroxyethyl-TPP<ref name="book">Voet, D., Voet, J.G., and Pratt, C.W. (2008). Fundamentals of biochemistry. Hoboken, NJ: John Wiley and Sons, Inc.</ref>. During this reaction, the catalytic Mg2+ ion coordinates octahedrally with three protein ligands: <scene name='Kenny_Coggins_Sandbox_1/Asp230_and_asn260/2'>Asp230 and Asn 260</scene>, which bind TPP, and <scene name='Kenny_Coggins_Sandbox_1/Glu262/2'>Gln262</scene><ref name="PMID" />. |
The second E1-catalyzed reaction is the transfer of the hydroxyethyl group to the lipoamide group of the next enzyme, dihydrolipoyltransacetylase (E2). The E2 lipoamide group consisting of lipoic acid linked to the amide group of a Lys residue. Lipoic acid contains a reactive cyclic disulfide that is reversibly reduced to give dihydrolipoamide. The ylide form of the hydroxyethyl group of the hydroxyethyl-TPP complex attacks this disulfide bond. TPP is then eliminated as it detaches with E1 and subsequently binds to the next pyruvate molecule<ref name="book" />. | The second E1-catalyzed reaction is the transfer of the hydroxyethyl group to the lipoamide group of the next enzyme, dihydrolipoyltransacetylase (E2). The E2 lipoamide group consisting of lipoic acid linked to the amide group of a Lys residue. Lipoic acid contains a reactive cyclic disulfide that is reversibly reduced to give dihydrolipoamide. The ylide form of the hydroxyethyl group of the hydroxyethyl-TPP complex attacks this disulfide bond. TPP is then eliminated as it detaches with E1 and subsequently binds to the next pyruvate molecule<ref name="book" />. | ||
Current revision
| |||||||||||
References
- ↑ Protein: Pyruvate dehydrogenase E1-beta, PdhB, C-terminal domain from Bacillus stearothermophilus. (2009). Retrieved from http://scop.mrc-lmb.cam.ac.uk
- ↑ Ciszak EM, Korotchkina LG, Dominiak PM, Sidhu S, Patel MS (June 2003). "Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase". J. Biol. Chem. 278 (23): 21240–6
- ↑ 3.0 3.1 Arjunan P, Nemeria N, Brunskill A, Chandrasekhar K, Sax M, Yan Y, Jordan F, Guest JR, Furey W. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution. Biochemistry. 2002 Apr 23;41(16):5213-21. PMID:11955070
- ↑ Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- ↑ 5.0 5.1 5.2 Voet, D., Voet, J.G., and Pratt, C.W. (2008). Fundamentals of biochemistry. Hoboken, NJ: John Wiley and Sons, Inc.
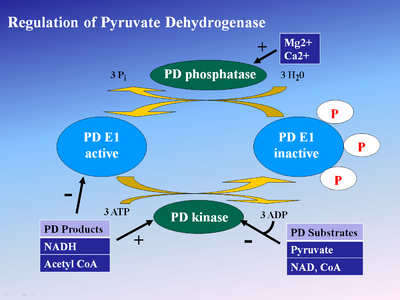
- ↑ Korotchkina LG, Patel MS. Probing the mechanism of inactivation of human pyruvate dehydrogenase by phosphorylation of three sites. J Biol Chem. 2001 Feb 23;276(8):5731-8. Epub 2000 Nov 22. PMID:11092882 doi:10.1074/jbc.M007558200
- ↑ Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci. 2007 Apr;64(7-8):830-49. PMID:17310282 doi:10.1007/s00018-007-6380-z
- ↑ Bisswanger, H. Substrate specificity of the pyruvate dehydrogenase complex from escherichia coli. J Biol Chem. 1981. Jan 25;256(2):815-822.
- ↑ Leung PS, Rossaro L, Davis PA, et al. (2007). "Antimitochondrial antibodies in acute liver failure: Implications for primary biliary cirrhosis". Hepatology 46 (5): 1436
- ↑ Frye, Richard E., and Paul J. Benke. "Pyruvate Dehydrogenase Complex Deficiency." EMedicine. 11 Dec. 2007. WebMD. 14 Dec. 2008