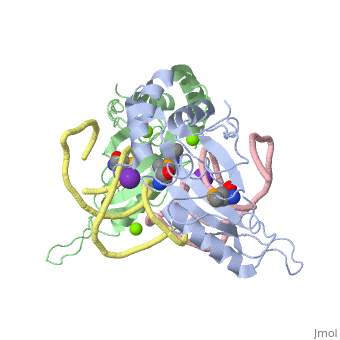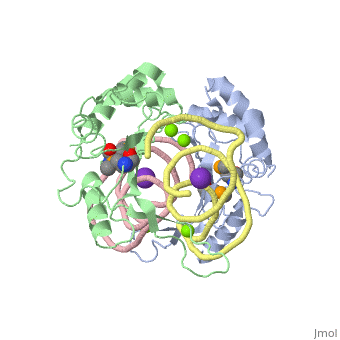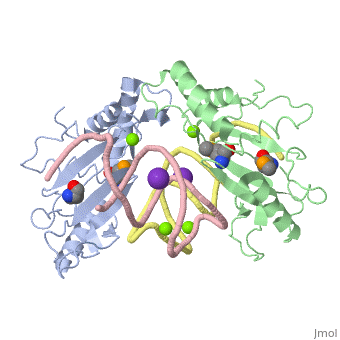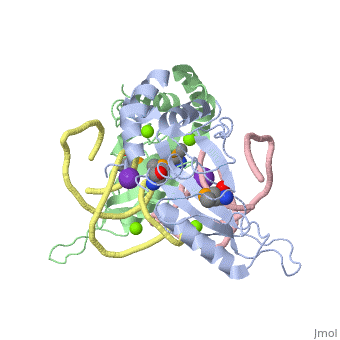
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1mji
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1mji' size='340' side='right'caption='[[1mji]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='1mji' size='340' side='right'caption='[[1mji]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1mji]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/ ] and [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1mji]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli] and [https://en.wikipedia.org/wiki/Thermus_thermophilus Thermus thermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1MJI OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1MJI FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=K:POTASSIUM+ION'>K</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=MSE:SELENOMETHIONINE'>MSE</scene></td></tr> |
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1mji FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1mji OCA], [https://pdbe.org/1mji PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1mji RCSB], [https://www.ebi.ac.uk/pdbsum/1mji PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1mji ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1mji FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1mji OCA], [https://pdbe.org/1mji PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1mji RCSB], [https://www.ebi.ac.uk/pdbsum/1mji PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1mji ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/RL5_THETH RL5_THETH] This is 1 of the proteins that binds and probably mediates the attachment of the 5S RNA into the large ribosomal subunit, where it forms part of the central protuberance. In the 70S ribosome it contacts protein S13 of the 30S subunit (forming bridge B1b) connecting the head of the 30S subunit to the top of the 50S subunit. The bridge itself contacts the P site tRNA and is implicated in movement during ribosome translocation. Also contacts the P site tRNA independently of the intersubunit bridge; the 5S rRNA and some of its associated proteins might help stabilize positioning of ribosome-bound tRNAs (By similarity).[HAMAP-Rule:MF_01333_B] | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 20: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1mji ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1mji ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The crystal structure of ribosomal protein L5 from Thermus thermophilus complexed with a 34-nt fragment comprising helix III and loop C of Escherichia coli 5S rRNA has been determined at 2.5 A resolution. The protein specifically interacts with the bulged nucleotides at the top of loop C of 5S rRNA. The rRNA and protein contact surfaces are strongly stabilized by intramolecular interactions. Charged and polar atoms forming the network of conserved intermolecular hydrogen bonds are located in two narrow planar parallel layers belonging to the protein and rRNA, respectively. The regions, including these atoms conserved in Bacteria and Archaea, can be considered an RNA-protein recognition module. Comparison of the T. thermophilus L5 structure in the RNA-bound form with the isolated Bacillus stearothermophilus L5 structure shows that the RNA-recognition module on the protein surface does not undergo significant changes upon RNA binding. In the crystal of the complex, the protein interacts with another RNA molecule in the asymmetric unit through the beta-sheet concave surface. This protein/RNA interface simulates the interaction of L5 with 23S rRNA observed in the Haloarcula marismortui 50S ribosomal subunit. | ||
| - | |||
| - | Detailed analysis of RNA-protein interactions within the bacterial ribosomal protein L5/5S rRNA complex.,Perederina A, Nevskaya N, Nikonov O, Nikulin A, Dumas P, Yao M, Tanaka I, Garber M, Gongadze G, Nikonov S RNA. 2002 Dec;8(12):1548-57. PMID:12515387<ref>PMID:12515387</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1mji" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Ribosomal protein L5|Ribosomal protein L5]] | *[[Ribosomal protein L5|Ribosomal protein L5]] | ||
| - | == References == | ||
| - | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Escherichia coli]] |
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Dumas | + | [[Category: Thermus thermophilus]] |
| - | [[Category: Garber | + | [[Category: Dumas P]] |
| - | [[Category: Gongadze | + | [[Category: Garber M]] |
| - | [[Category: Nevskaya | + | [[Category: Gongadze G]] |
| - | [[Category: Nikonov | + | [[Category: Nevskaya N]] |
| - | [[Category: Nikonov | + | [[Category: Nikonov O]] |
| - | [[Category: Nikulin | + | [[Category: Nikonov S]] |
| - | [[Category: Perederina | + | [[Category: Nikulin A]] |
| - | [[Category: Tanaka | + | [[Category: Perederina A]] |
| - | [[Category: Yao | + | [[Category: Tanaka I]] |
| - | + | [[Category: Yao M]] | |
| - | + | ||
Revision as of 08:37, 10 April 2024
DETAILED ANALYSIS OF RNA-PROTEIN INTERACTIONS WITHIN THE BACTERIAL RIBOSOMAL PROTEIN L5/5S RRNA COMPLEX
| |||||||||||
Categories: Escherichia coli | Large Structures | Thermus thermophilus | Dumas P | Garber M | Gongadze G | Nevskaya N | Nikonov O | Nikonov S | Nikulin A | Perederina A | Tanaka I | Yao M