User:Eric Martz/AlphaFold3 case studies
From Proteopedia
(→8JRP Chain A: Partially Untemplated) |
(→8JRP Chain A: Partially Untemplated) |
||
| Line 44: | Line 44: | ||
[https://alphafoldserver.com AlphaFold3 Server] FAQ says that when [[empirical models|empirical]] templates exist in the [[wwPDB]], they will be used unconditionally, adding that “… the server searches PDB for template structures with a cutoff date of 30th September 2021 …”. | [https://alphafoldserver.com AlphaFold3 Server] FAQ says that when [[empirical models|empirical]] templates exist in the [[wwPDB]], they will be used unconditionally, adding that “… the server searches PDB for template structures with a cutoff date of 30th September 2021 …”. | ||
| - | According to this, the N-terminal half of chain A of [[8jrp]]<ref name="choice">[[8jrp]] was chosen despite its poor resolution of 3.6 Å because the better-resolution [[8jrn]], 2.6 Å, is not available in PDB format. A rigid superposition by FATCAT matched all 689 residues with coordinates, giving [[RMSD]] 0.7 Å, a near perfect match.</ref> (120-519) should be '''untemplated''', since [https://www.rcsb.org/search?request=%7B%22query%22%3A%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22evalue_cutoff%22%3A0.1%2C%22identity_cutoff%22%3A0%2C%22sequence_type%22%3A%22protein%22%2C%22value%22%3A%22IDFKDVTYLTEEKVYEILELCREREDYSPLIRVIGRVFSSAEALVQSFRKVKQHTKEELKSLQAKDEDKDEDEKEKAACSAAAMEEDSEASSSRIGDSSQGDNNLQKLGPDDVSVDIDAIRRVYTRLLSNEKIETAFLNALVYLSPNVECDLTYHNVYSRDPNYLNLFIIVMENRNLHSPEYLEMALPLFCKAMSKLPLAAQGKLIRLWSKYNADQIRRMMETFQQLITYKVISNEFNSRNLVNDDDAIVAASKCLKMVYYANVVGGEVDTNHNEEDDEEPIPESSELTLQELLGEERRNKKGPRVDPLETELGVKTLDCRKPLIPFEEFINEPLNEVLEMDKDYTFFKVETENKFSFMTCPFILNAVTKNLGLYYDNRIRMYSERRITVLYSLVQGQ%22%7D%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%2C%22return_type%22%3A%22polymer_entity%22%2C%22request_options%22%3A%7B%22paginate%22%3A%7B%22start%22%3A0%2C%22rows%22%3A25%7D%2C%22results_content_type%22%3A%5B%22experimental%22%5D%2C%22sort%22%3A%5B%7B%22sort_by%22%3A%22score%22%2C%22direction%22%3A%22desc%22%7D%5D%2C%22scoring_strategy%22%3A%22combined%22%7D%2C%22request_info%22%3A%7B%22query_id%22%3A%223e8b3e387c871e8978a2e764d8109fbd%22%7D%7D a sequence search] finds no matching entries prior to 2023. Being untemplated should make structure prediction more challenging. Of course, a structure need not be sequence-identical to serve as a template. A search for similar chain structures finds no good matches (top hits "not significantly similar" according to FATCAT), consistent with this N-terminal half being untemplated. The C-terminal half (520-869) is templated by 2 entries, e.g. [[1c4z]]<ref name="1c4z">A FATCAT rigid superposition of 8jrp chain A with 1cz4 chain C shows a twist between the two domains. A FATCAT flexible superposition with one twist gives RMSD 1.3 Å for all 350 alpha carbons.</ref>, a 1999 X-ray 2.6 Å resolution structure. | + | According to this, the N-terminal half of chain A of [[8jrp]]<ref name="choice">[[8jrp]] was chosen despite its poor resolution of 3.6 Å because the better-resolution [[8jrn]], 2.6 Å, is not available in PDB format. A rigid superposition by FATCAT matched all 689 residues with coordinates, giving [[RMSD]] 0.7 Å, a near perfect match.</ref> (120-519) should be '''untemplated''', since [https://www.rcsb.org/search?request=%7B%22query%22%3A%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22evalue_cutoff%22%3A0.1%2C%22identity_cutoff%22%3A0%2C%22sequence_type%22%3A%22protein%22%2C%22value%22%3A%22IDFKDVTYLTEEKVYEILELCREREDYSPLIRVIGRVFSSAEALVQSFRKVKQHTKEELKSLQAKDEDKDEDEKEKAACSAAAMEEDSEASSSRIGDSSQGDNNLQKLGPDDVSVDIDAIRRVYTRLLSNEKIETAFLNALVYLSPNVECDLTYHNVYSRDPNYLNLFIIVMENRNLHSPEYLEMALPLFCKAMSKLPLAAQGKLIRLWSKYNADQIRRMMETFQQLITYKVISNEFNSRNLVNDDDAIVAASKCLKMVYYANVVGGEVDTNHNEEDDEEPIPESSELTLQELLGEERRNKKGPRVDPLETELGVKTLDCRKPLIPFEEFINEPLNEVLEMDKDYTFFKVETENKFSFMTCPFILNAVTKNLGLYYDNRIRMYSERRITVLYSLVQGQ%22%7D%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%2C%22return_type%22%3A%22polymer_entity%22%2C%22request_options%22%3A%7B%22paginate%22%3A%7B%22start%22%3A0%2C%22rows%22%3A25%7D%2C%22results_content_type%22%3A%5B%22experimental%22%5D%2C%22sort%22%3A%5B%7B%22sort_by%22%3A%22score%22%2C%22direction%22%3A%22desc%22%7D%5D%2C%22scoring_strategy%22%3A%22combined%22%7D%2C%22request_info%22%3A%7B%22query_id%22%3A%223e8b3e387c871e8978a2e764d8109fbd%22%7D%7D a sequence search] finds no matching entries prior to 2023. Being untemplated should make structure prediction more challenging. Of course, a structure need not be sequence-identical to serve as a template. A search for similar chain structures finds no good matches (top hits "not significantly similar" according to FATCAT), consistent with this N-terminal half being untemplated. The C-terminal half (520-869) is templated by 2 entries, e.g. [[1c4z]]<ref name="1c4z">A FATCAT rigid superposition of 8jrp chain A with 1cz4 chain C shows a twist between the two domains. A FATCAT flexible superposition with one twist gives RMSD 1.3 Å for all 350 alpha carbons. Thus, 1c4z would be an excellent template for predicting the C-terminal half of 8jrp chain A.</ref>, a 1999 X-ray 2.6 Å resolution structure. |
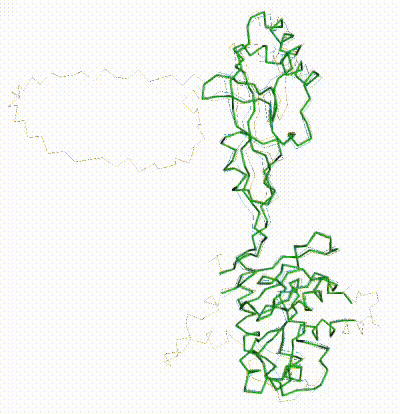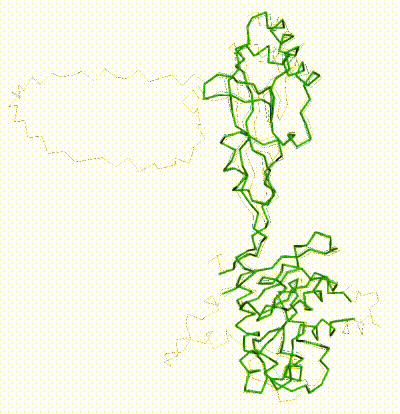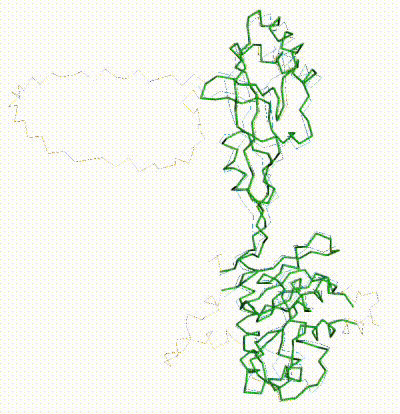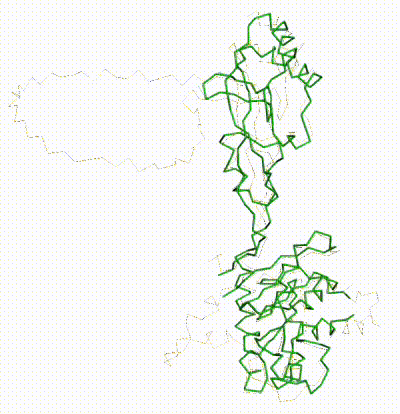
Chain R in [[8gcr]] is human ubiquitin-protein ligase E3A. The full-length sequence, [https://www.uniprot.org/uniprotkb/Q05086/entry Q05086], has 875 amino acids. The 3.4 Å resolution [[cryo-EM]] structure indicates that the full-length protein was imaged, but each end of the model lacks coordinates for over 100 residues. The ends of the model are sequence numbers 126 and 761 (length 636), but three loops (1, 2, 3) of lengths 62, 7 and 6 respectively are missing coordinates, leaving 561 amino acids with coordinates. Conveniently, the sequence numbers in UniProt and [[8gcr]] are the same. | Chain R in [[8gcr]] is human ubiquitin-protein ligase E3A. The full-length sequence, [https://www.uniprot.org/uniprotkb/Q05086/entry Q05086], has 875 amino acids. The 3.4 Å resolution [[cryo-EM]] structure indicates that the full-length protein was imaged, but each end of the model lacks coordinates for over 100 residues. The ends of the model are sequence numbers 126 and 761 (length 636), but three loops (1, 2, 3) of lengths 62, 7 and 6 respectively are missing coordinates, leaving 561 amino acids with coordinates. Conveniently, the sequence numbers in UniProt and [[8gcr]] are the same. | ||
Revision as of 21:38, 14 November 2024
|
This page is under construction. This notice will be removed when it is completed. Eric Martz 02:03, 13 November 2024 (UTC) |
The following case studies of predictions by the AlphaFold3 Server were done in November 2024. At that time, the AlphaFold Database had been generated with AlphaFold2. Superpositions and RMSD values were obtained from FATCAT. Rocking animations were generated by FirstGlance in Jmol, and morphs were captured from FATCAT.
Contents |
AlphaFold3 Example 8AW3
Summary: This is one of three examples featured in the AlphaFold3 Server. AlphaFold3 predicted the fold of the mid-341 residues of tRNA A34 deaminase (UniProt Q381Q7) accurately, when compared to the empirical model in 8aw3. It made no confident predictions for what appear to be 90 residues in two disordered loops, and a 25-residue disordered C terminus. In contrast, the prediction from the AlphaFold2 Database was bent relative to the empirical model, but otherwise largely accurate.
UniProt Q381Q7 (tRNA A34 deaminase) is the longest chain in the example 8aw3 provided by the AlphaFold3 Server. In 8aw3, its chain name is "3". 8aw3 is a cryo-EM structure with resolution 3.6 Å. 8aw3 indicates that the experimental material included full-length chain 3 of 369 residues, but 28 end-residues lack coordinates, so the coordinates run from UniProt 4-344, length 341. Within this range, there are two loops lacking coordinates of lengths 49 and 13, leaving 279 amino acids with coordinates. Neither missing loop is predicted to be disordered at UniProt, although the estimated disorder propensity is high for both.
Q381Q7: AlphaFold3 vs Empirical Structure
The overall 4-chain prediction (for the complex in 8aw3) has pTM of 0.75, at the low end of the range 0.7-0.9 deemed "confident" by the server. Both of the missing loops have "no-confidence" (red) coordinates. The longer one is modeled as a large circle protruding from the surface of the protein, typical for loops with intrinsic disorder. The average pLDDT for chain 3 is 68 (weak confidence), but with the large disordered loop removed, this increases to 74[1].
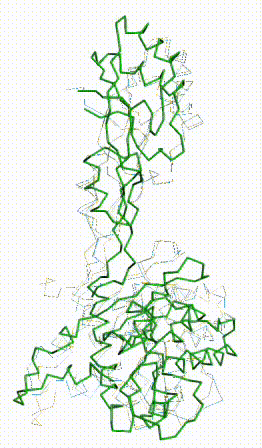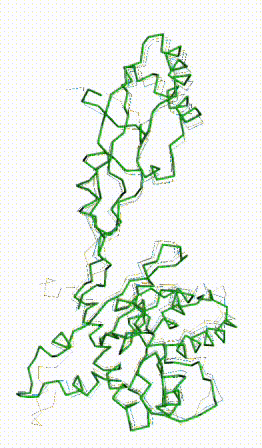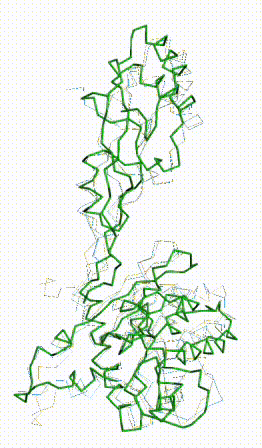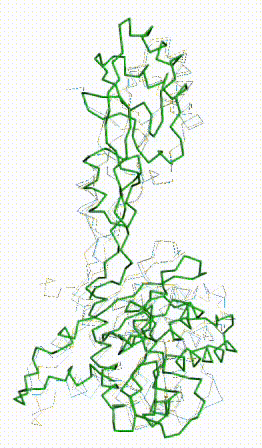
The AlphaFold3 prediction superposes (FATCAT rigid) with all 279 alpha carbons of chain 3 of 8aw3 with RMSD 2.2 Å. The morph shows the largest discrepancy in the red (no confidence) loop at center bottom, which has sequence SNSGCRKSNR (238-247 in the numbering of both UniProt and 8aw3). These residues have coordinates in 8aw3, but 3 residues, CRK, have incomplete sidechains.
|
AlphaFold3 prediction for Q38107. |
FATCAT morph between AlphaFold3 prediction and chain 3 of 8aw3. |
Q381Q7: AlphaFold3 vs AlphaFold2 Database
|
The long disordered loop was in different positions in the AlphaFold3 vs. AlphaFold2 Database models. When it was included, FATCAT superposed it between models, causing the RMSD to be 4.8-4.9 Å (rigid or flexible superposition). Deleting that loop from the models resulted in a much better flexible superposition RMSD value of 2.0 Å for 312 alpha carbons (98% of the 317 present). This close superposition required flexibility. The rigid FATCAT superposition still had RMSD 4.5 Å. The morph shows bending and twisting between the two models. Since the AlphaFold2 Database prediction is bent/twisted relative to the AlphaFold3 prediction, and since the latter superposes well with the empirical model in a rigid superposition, the AlphaFold2 Database prediction should not superpose as well with the empirical model in a rigid superposition. Indeed, the rigid superposition has RMSD 4.1 Å for 270 (of 279) alpha carbons (morph not shown). (Flexible superposition achieved RMSD 2.4 Å.) | |
|
FATCAT morph between AlphaFold3 prediction and AlphaFold2 Database prediction for Q381Q7. |
8JRP Chain A: Partially Untemplated
AlphaFold3 Server FAQ says that when empirical templates exist in the wwPDB, they will be used unconditionally, adding that “… the server searches PDB for template structures with a cutoff date of 30th September 2021 …”.
According to this, the N-terminal half of chain A of 8jrp[2] (120-519) should be untemplated, since a sequence search finds no matching entries prior to 2023. Being untemplated should make structure prediction more challenging. Of course, a structure need not be sequence-identical to serve as a template. A search for similar chain structures finds no good matches (top hits "not significantly similar" according to FATCAT), consistent with this N-terminal half being untemplated. The C-terminal half (520-869) is templated by 2 entries, e.g. 1c4z[3], a 1999 X-ray 2.6 Å resolution structure.
Chain R in 8gcr is human ubiquitin-protein ligase E3A. The full-length sequence, Q05086, has 875 amino acids. The 3.4 Å resolution cryo-EM structure indicates that the full-length protein was imaged, but each end of the model lacks coordinates for over 100 residues. The ends of the model are sequence numbers 126 and 761 (length 636), but three loops (1, 2, 3) of lengths 62, 7 and 6 respectively are missing coordinates, leaving 561 amino acids with coordinates. Conveniently, the sequence numbers in UniProt and 8gcr are the same.
Loops missing/disordered
Loops 1 and 2, missing from 8gcr, are predicted to be disordered at RCSB. Missing loop 3 (387-393) is not. Sequence range 394-424 is predicted to be disordered, but has coordinates in 8gcr.
AlphaFold 3 prediction
|
morph | ||
|
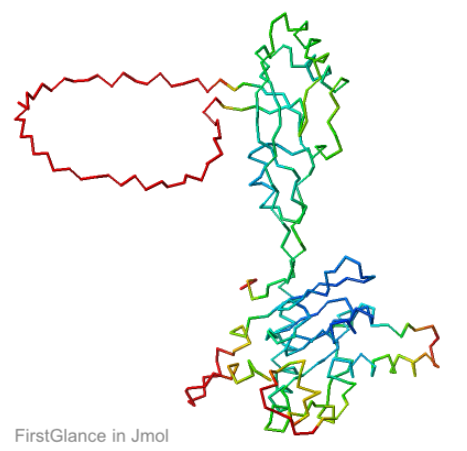
Chain R of 8gcr. Missing ends and loops shown as "empty baskets" by FirstGlance in Jmol.
|
AlphaFold3 prediction for Q05086. |
FATCAT morph between AlphaFold3 prediction and chain R of 8gcr. |
See Also
- AlphaFold3 Server alphafoldserver.com
- How to predict structures with AlphaFold
- AlphaFold/Index lists pages in Proteopedia that relate to AlphaFold.
Notes
- ↑ Average pLDDT values were obtained with FirstGlance in Jmol, which reports the minimum, average, and maximum in the upper right as "Reliability".
- ↑ 8jrp was chosen despite its poor resolution of 3.6 Å because the better-resolution 8jrn, 2.6 Å, is not available in PDB format. A rigid superposition by FATCAT matched all 689 residues with coordinates, giving RMSD 0.7 Å, a near perfect match.
- ↑ A FATCAT rigid superposition of 8jrp chain A with 1cz4 chain C shows a twist between the two domains. A FATCAT flexible superposition with one twist gives RMSD 1.3 Å for all 350 alpha carbons. Thus, 1c4z would be an excellent template for predicting the C-terminal half of 8jrp chain A.