Sandbox Reserved 1847
From Proteopedia
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
===What are the SARS-CoV2 Minibinders?=== | ===What are the SARS-CoV2 Minibinders?=== | ||
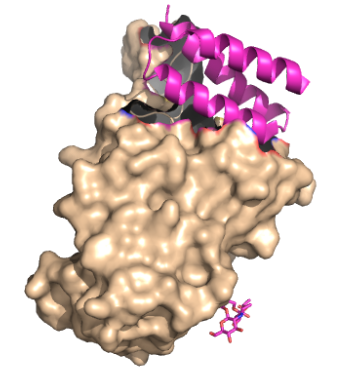
| - | These mini proteins target the interaction between ACE2 and | + | These mini proteins target the interaction between ACE2 and SARS-CoV-2 spike protein <ref name="Longxing">PMID:32907861</ref>. The mini binders are small proteins designed to bind to the SARS-CoV-2 spike protein with a greater affinity than ACE2 <ref name="Longxing">PMID:32907861</ref>. These mini binders reduce the viral burden of SARS-CoV-2 in mice <ref name="Case">PMID:34192518</ref>. Minibinders were de novo ITALICS designed to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein <ref name="Longxing">PMID:32907861</ref>. The binding region between <scene name='10/1075251/Ace2_and_rbd/3'>spike protein and ACE2</scene> (PDB: [https://www.rcsb.org/structure/8K4U 8K4U]) makes it seem like binding region is telling how these proteins were designed. Taking a closer look at the SARS-CoV-2 disease pathway shows where the minibinders target the interaction between ACE2 and SARS-CoV-2 spike protein. |
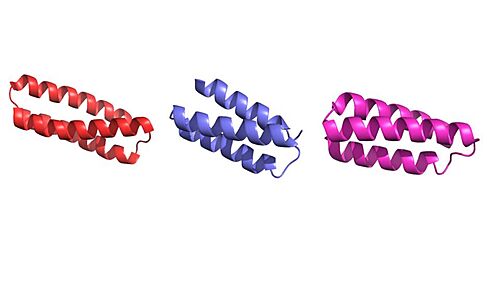
[[Image:Untitled presentation.jpg|500 px|right|thumb|Figure 1. Image of the individual helices of AHB2, LCB1, and LCB3, respectively.]] | [[Image:Untitled presentation.jpg|500 px|right|thumb|Figure 1. Image of the individual helices of AHB2, LCB1, and LCB3, respectively.]] | ||
Revision as of 16:50, 27 April 2025
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Contents |
Structure
Introduction
What are the SARS-CoV2 Minibinders?
These mini proteins target the interaction between ACE2 and SARS-CoV-2 spike protein [1]. The mini binders are small proteins designed to bind to the SARS-CoV-2 spike protein with a greater affinity than ACE2 [1]. These mini binders reduce the viral burden of SARS-CoV-2 in mice [2]. Minibinders were de novo ITALICS designed to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein [1]. The binding region between (PDB: 8K4U) makes it seem like binding region is telling how these proteins were designed. Taking a closer look at the SARS-CoV-2 disease pathway shows where the minibinders target the interaction between ACE2 and SARS-CoV-2 spike protein.
COVID-19 Disease Pathway
Understanding the pathway of the COVID-19 virus is essential to understanding the mechanism in which the virus’ surface proteins attach to the mini binders. The COVID-19 virus has spike proteins on its surface that bind to the host cell receptor, known as ACE2, and this allows the virus to remain anchored to the host for viral entry [3]. When the spike protein binds to the receptor, ACE2 for example, the cell membrane-associated protease, protease serine 2 TMPRSS2 promotes viral entry by activating the spike protein [4]. The activated spike protein is able to cleave itself into S1 and S2 subunits [4]. The S2 subunit is in charge of viral entry and does this through conformational changes [4]. The S2 subunit will insert it's FP domain into the host cell's membrane, and this will trigger an interaction with the HR2 domain and HR1 trimer to form the 6-helical bundle to bring the viral envelope and cell membrane in close enough distance for viral fusion and ultimately viral entry [4]. Once the virus is within the host cell, it is able to translate viral proteins, eliciting an immune response and spreading the viral particles throughout the body [4].
COVID-19 Viral Infection Interruption
The primary goal of the mini binders is to prevent the spike proteins from binding to ACE2, and when the mini binders are bound to the spike protein, the virus is unable to anchor itself to the host protein [1]. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they are able to effectively prevent the entry of the virus and ultimately prevent an immune response [1]. Targeting this specific interaction between the COVID-19 spike protein has proven effective and is hopeful target for future therapeutics to treat the virus [4]. LCB1 proved to be quite effective at weakening the immune response, compared to the other mini binders, which can be explained by the binding interface between the spike protein and LCB1 [1].
Expectations of this page
This page will dive into the method of mini binders effectively preventing the entry of the viral SARS-CoV-2 spike protein to enter a host cell. Taking a closer look at the structure of the SARS-CoV-2 spike protein, this will provide a deeper understanding of how it binds to host a cell membrane (ACE2) and give context to how the mini binders are capable of binding to it. Understanding the ACE2 receptor on host cells will give an expectation for how mini binders will bind to the spike protein, stealing the parts of the structure of ACE2 to create a protein with a greater binding affinity to outcompete ACE2.
Laying the foundation for the mini binders, we will then take a look at how the mini binders are designed to obtain the best possible helical structure. With that, then it is finally time to look at the receptor binding domain between the various mini binders and their interactions with the spike protein.
Design
The binding region between show how these minibinders were designed. These minibinders, and , were designed de novo to mimic the binding of ACE2 to spike protein [1]. Using Rotamer Interaction Field (RIF) docking, the proteins were able to make the most efficient bonding using the ACE2 spike protein binding interface [1]. Using Site Saturation Mutagenesis (SSM), every residue in the minibinder’s helix scaffold will be substituted with each of the 20 amino acids, one at a time [5]. Forming SSM libraries, each of the libraries converged on a small number of closely related sequences, and from these libraries, the design was selected for AHB2 and LCB1 to find the sequence that yields a protein with a high affinity for the spike proteins receptor binding domain [1]. AHB2 was designed using ACE2 helix scaffold, while LCB1 and LCB3 were designed full from scratch, attempting to make the best possible helix with the greatest affinity for the spike protein receptors [1]. Although LCB1 was designed before LCB3, LCB3 was less effective at neutralizing the viral response with a higher IC50 value [1].
| |||||||||||
|