Sand box 326
From Proteopedia
| Line 22: | Line 22: | ||
| - | == Sequence Analysis | + | == Sequence Analysis == |
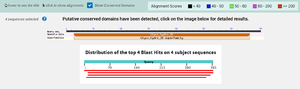
According to BLAST, 3CBW has similar sequences to 2QHA (Beta-1,4-mannanase) and 2WHK (Mannan endo-1,4-beta-mannosidase). This means that 3CBW is in the mannanase super family. | According to BLAST, 3CBW has similar sequences to 2QHA (Beta-1,4-mannanase) and 2WHK (Mannan endo-1,4-beta-mannosidase). This means that 3CBW is in the mannanase super family. | ||
| Line 28: | Line 28: | ||
[[Image:BLAST.png |300px|center|thumb|]] | [[Image:BLAST.png |300px|center|thumb|]] | ||
| - | == Structural Analysis | + | == Structural Analysis == |
Using Right-hand SPRITE analysis, it showed that 3BCW had similar residues to small amino acid chains. Specific residues are Ala. 76, Ile. 79, Gly. 61, Val. 293, the first two being on the A chain side and the second being on the B chain side of the 3CBW. Comparing 3CBW to 1BHG, which is human beta-glucuronidase, the RMSD value was a difference of 0.46 angstroms. | Using Right-hand SPRITE analysis, it showed that 3BCW had similar residues to small amino acid chains. Specific residues are Ala. 76, Ile. 79, Gly. 61, Val. 293, the first two being on the A chain side and the second being on the B chain side of the 3CBW. Comparing 3CBW to 1BHG, which is human beta-glucuronidase, the RMSD value was a difference of 0.46 angstroms. | ||
| Line 37: | Line 37: | ||
| - | == Substrates | + | == Substrates == |
Using SwissDock, it was determined that larger, cyclic molecules are good substrates that would have the best binding affinity to 3CBW, each had about a -7 kcal/mol binding affinity. These large molecules are 4-Nitrophenyl N-acetyl-β-D-glucosaminide, 4-Nitrophenyl α-D-glucopyranoside, and p-nitrophenyl phosphate. | Using SwissDock, it was determined that larger, cyclic molecules are good substrates that would have the best binding affinity to 3CBW, each had about a -7 kcal/mol binding affinity. These large molecules are 4-Nitrophenyl N-acetyl-β-D-glucosaminide, 4-Nitrophenyl α-D-glucopyranoside, and p-nitrophenyl phosphate. | ||
| - | == Theoretical Functionality and Proposed Bodily Purpose | + | == Theoretical Functionality and Proposed Bodily Purpose == |
| - | According to our research the likely | + | According to our research, 3CBW is a glycoside hydrolase whose job is to hydrolyze glycosidic bonds between two or more carbohydrates. Since this protein was found to most likely come from the bacterial species Bacillus Subtilis, it is highly likely that this protein was found in the human gastrointestinal tract. That is because Bacillus Subtilis is a probiotic bacteria and can aid in digestion and support a healthy microbiome. |
| + | |||
| + | It offers several potential gut health and immunity benefits, and it can be sourced through probiotic supplements or by eating fermented foods. | ||
| + | |||
| + | Substrates: 4-Nitrophenyl N-acetyl-β-D-glucosaminide, 4-Nitrophenyl α-D-glucopyranoside, and P-Nitrophenyl Phosphate | ||
| + | |||
| + | Binding sites: | ||
</StructureSection> | </StructureSection> | ||
| - | == References | + | == References == |
A) Dhawan, S.; Kaur, J.; Microbial Mannanases: An Overview of Production and Applications. Critical Reviews in Biotechnology 2007, 27, 197-216. DOI: 10.1080/07388550701775919 | A) Dhawan, S.; Kaur, J.; Microbial Mannanases: An Overview of Production and Applications. Critical Reviews in Biotechnology 2007, 27, 197-216. DOI: 10.1080/07388550701775919 | ||
Revision as of 12:55, 28 April 2025
3CBW Structure and Proposed Functionality
(NOTE TO ALL EDITORS: This page is part of a final project for a biochemistry lab at Elizabethtown College. Please do not edit this.)
3CBW is a homodimeric protein complex that originates from the bacterial species Bacillus Subtilis and has a mass of 80.65 kDa. It is a member of the Glycoside Hydrolase super family with structural and sequential similarities to hydrolases and mannanases. Current evidence suggests it causes the hydrolysis of glycosidic linkages via a LEU. 176 or ILE. 275 active site.
| |||||||||||
References
A) Dhawan, S.; Kaur, J.; Microbial Mannanases: An Overview of Production and Applications. Critical Reviews in Biotechnology 2007, 27, 197-216. DOI: 10.1080/07388550701775919 B) Soni, H.; Rawat, H. K.; Pletschke, B. I.; Kango, N. Purification and characterization of Beta-mannanase from Aspergillus terreus and its applicability in depolymerization of mannans and saccharification of lignocellulosic biomass. Biotech 2016, 6, 136. DOI: 10.1007/s13205-016-0454-2 C) Cheng, L.; Duan, S.; Feng, X.; Zheng, K.; Yang, Q.; Liu, Z. Purification and Characterization of a Thermostable Beta-Mannanase from Bacillus subtilis BE-91: Potential Application in Inflammatory Diseases. BioMed Research International 2016, 2016, 1-7. DOI: 10.1155/2016/6380147