We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox: 5VKQ
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
The NOMPC (No mechanoreceptor potential C) is a mechanosensitive ion channel essential for hearing, touch and locomotion ([https://en.wikipedia.org/wiki/Proprioception proprioception]) in ''Drosophila melanogaster''. It responds directly to mechanical stimuli such as pressure and stretch, converting physical forces into electrical signals. NOMPC is classified as a [https://en.wikipedia.org/wiki/Transient_receptor_potential_channel transient receptor potential] (TRP) channel, a superfamily of membrane bound proteins involved with [https://en.wikipedia.org/wiki/Signal_transduction stimulus detection and sensory transduction] in various animal cells. When first discovered, NOMPC introduced a novel class of receptors, the TRPN. Although it was first described in ''Drosophila melanogaster'' <ref>PMID:10744543</ref>, [https://en.wikipedia.org/wiki/Sequence_homology homologs] of NOMPC were found in other animals, including the nematode worm ''Caenorhabditis elegans'' and the zebrafish. | The NOMPC (No mechanoreceptor potential C) is a mechanosensitive ion channel essential for hearing, touch and locomotion ([https://en.wikipedia.org/wiki/Proprioception proprioception]) in ''Drosophila melanogaster''. It responds directly to mechanical stimuli such as pressure and stretch, converting physical forces into electrical signals. NOMPC is classified as a [https://en.wikipedia.org/wiki/Transient_receptor_potential_channel transient receptor potential] (TRP) channel, a superfamily of membrane bound proteins involved with [https://en.wikipedia.org/wiki/Signal_transduction stimulus detection and sensory transduction] in various animal cells. When first discovered, NOMPC introduced a novel class of receptors, the TRPN. Although it was first described in ''Drosophila melanogaster'' <ref>PMID:10744543</ref>, [https://en.wikipedia.org/wiki/Sequence_homology homologs] of NOMPC were found in other animals, including the nematode worm ''Caenorhabditis elegans'' and the zebrafish. | ||
| - | (Click to reset | + | <scene name='10/1083740/Homotetramer_0/1'>(Click to reset the protein view.)</scene> |
== Structure and mechanism of action == | == Structure and mechanism of action == | ||
| Line 13: | Line 13: | ||
NOMPC structure was first resolved in 2017 using [https://proteopedia.org/wiki/index.php/Cryo-EM Cryo-EM] technique <ref name="jin2016">PMID:28658211</ref>, with a resolution of 3.6 Å. At this resolution, models can distinguish between atoms. This level of detail is essential for understanding the mechanism of action of this protein. | NOMPC structure was first resolved in 2017 using [https://proteopedia.org/wiki/index.php/Cryo-EM Cryo-EM] technique <ref name="jin2016">PMID:28658211</ref>, with a resolution of 3.6 Å. At this resolution, models can distinguish between atoms. This level of detail is essential for understanding the mechanism of action of this protein. | ||
| - | NOMPC is a [https://en.wikipedia.org/wiki/Tetrameric_protein#Homotetramers_and_heterotetramers homotetramer] (click to highlight each monomer), meaning its biologically functional arrangement is formed by the [https://proteopedia.org/wiki/index.php/Non-covalent_interactions non-covalent] interaction between four equal subunits, i.e. monomers. The monomers interact with each other through domain-swap interactions, where each monomer contributes part of its structure to stabilize its neighbors <ref>PMID:27867057</ref>, leading to a stable, ring-like assembly. Effectively, the monomers interlock. (Click here to see a single monomer.) | + | NOMPC is a [https://en.wikipedia.org/wiki/Tetrameric_protein#Homotetramers_and_heterotetramers homotetramer] <scene name='10/1083740/Homotetramer_1/1'>(click to highlight each monomer)</scene>, meaning its biologically functional arrangement is formed by the [https://proteopedia.org/wiki/index.php/Non-covalent_interactions non-covalent] interaction between four equal subunits, i.e. monomers. The monomers interact with each other through domain-swap interactions, where each monomer contributes part of its structure to stabilize its neighbors <ref>PMID:27867057</ref>, leading to a stable, ring-like assembly. Effectively, the monomers interlock. <scene name='10/1083740/Monomer_0/1'>(Click here to see a single monomer.)</scene> |
| - | By resolving its structure, researchers highlighted the presence of [https://en.wikipedia.org/wiki/Ankyrin_repeat 29 ankyrin repeats] per monomer, at the N-terminus. (Click here to highlight a single ankyrin repeat.) This is the largest number among TRP channels. An ankyrin repeat is a [https://en.wikipedia.org/wiki/Structural_motif motif] of about 33 amino acids repeated in tandem (i.e., one behind the other). Each repeat adopts a helix-turn-helix conformation. Ankyrin repeats are known to mediate [https://en.wikipedia.org/wiki/Protein%E2%80%93protein_interaction protein-protein interactions] <ref>InterPro (IPR002110). Ankyrin repeat. Available at: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR002110/. Accessed on 2025 June 19.</ref><ref>PMID:17176038</ref>. (Click here to highlight the ankyrin repeat domain.) | ||
| - | As other TRP channels, NOMPC has a transmembrane domain ( | + | By resolving its structure, researchers highlighted the presence of [https://en.wikipedia.org/wiki/Ankyrin_repeat 29 ankyrin repeats] per monomer, at the N-terminus. <scene name='10/1083740/Monomer_1_singlear/1'>(Click here to highlight a single ankyrin repeat.)</scene> This is the largest number among TRP channels. An ankyrin repeat is a [https://en.wikipedia.org/wiki/Structural_motif motif] of about 33 amino acids repeated in tandem (i.e., one behind the other). Each repeat adopts a helix-turn-helix conformation. Ankyrin repeats are known to mediate [https://en.wikipedia.org/wiki/Protein%E2%80%93protein_interaction protein-protein interactions] <ref>InterPro (IPR002110). Ankyrin repeat. Available at: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR002110/. Accessed on 2025 June 19.</ref><ref>PMID:17176038</ref>. <scene name='10/1083740/Monomer_1_ardomain/1'>(Click here to highlight the ankyrin repeat domain.)</scene> |
| + | |||
| + | As other TRP channels, NOMPC has a <scene name='10/1083740/Monomer_1_ar_tm/1'>transmembrane domain</scene> (blue) characterized by a series of six alpha-helices (S1–S6). Additional <scene name='10/1083740/Monomer_1_ar_tm_lh/1'>linker helices </scene> (orange) connect the ankyrin repeat domain and the transmembrane domain. Following the C-terminus portion, there is an intracellular <scene name='10/1083740/Monomer_1_ar_tm_lh_trp/1'>TRP domain</scene> (magenta). The rest of the C-terminus is mostly unstructured. | ||
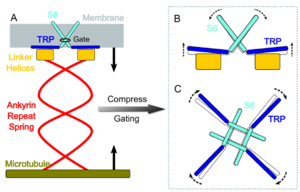
Although C-terminal, the TRP domain is “sandwiched” between the linker helices and the transmembrane portion, when looking at NOMPC tertiary structure (see Figure 2 and the 3D visualization). This positioning is important since the TRP domain movement regulates the pore formation (see Figure 1) <ref>PMID:34101577</ref>. | Although C-terminal, the TRP domain is “sandwiched” between the linker helices and the transmembrane portion, when looking at NOMPC tertiary structure (see Figure 2 and the 3D visualization). This positioning is important since the TRP domain movement regulates the pore formation (see Figure 1) <ref>PMID:34101577</ref>. | ||
| Line 23: | Line 24: | ||
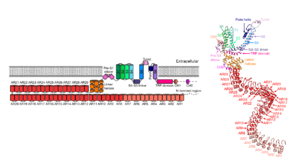
[[Image:5vkq_proteopedia_fig2.png|300px|right|thumb|''Figure 2.'' Monomeric representation of NOMPC. Left: two-dimensional schematic. Right: tertiary structure of NOMPC modelled with Cryo-EM data. (Source: Jin et al. Nature. 2017;547(7661):118-122. doi:10.1038/nature22981)]] | [[Image:5vkq_proteopedia_fig2.png|300px|right|thumb|''Figure 2.'' Monomeric representation of NOMPC. Left: two-dimensional schematic. Right: tertiary structure of NOMPC modelled with Cryo-EM data. (Source: Jin et al. Nature. 2017;547(7661):118-122. doi:10.1038/nature22981)]] | ||
| - | (Click to reset to the tetramer view.) | + | <scene name='10/1083740/Homotetramer_1/1'>(Click to reset to the tetramer colored view.)</scene> |
NOMPC gating is thought to be triggered by tethering of its [https://en.wikipedia.org/wiki/Protein_domain ankyrin repeat domain] to [https://en.wikipedia.org/wiki/Microtubule microtubules] of the [https://en.wikipedia.org/wiki/Cytoskeleton cytoskeleton]. Structural modelling of this protein suggests that the ankyrin repeat domain in each monomer forms a helical spring. This structure suggests an interaction between the NOMPC and the cytoskeleton, and could suggest the opening of the ion channel in response to cytoskeleton displacement. | NOMPC gating is thought to be triggered by tethering of its [https://en.wikipedia.org/wiki/Protein_domain ankyrin repeat domain] to [https://en.wikipedia.org/wiki/Microtubule microtubules] of the [https://en.wikipedia.org/wiki/Cytoskeleton cytoskeleton]. Structural modelling of this protein suggests that the ankyrin repeat domain in each monomer forms a helical spring. This structure suggests an interaction between the NOMPC and the cytoskeleton, and could suggest the opening of the ion channel in response to cytoskeleton displacement. | ||
Revision as of 20:44, 22 June 2025
| |||||||||||
References
- ↑ Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000 Mar 24;287(5461):2229-34. PMID:10744543 doi:10.1126/science.287.5461.2229
- ↑ 2.0 2.1 Jin P, Bulkley D, Guo Y, Zhang W, Guo Z, Huynh W, Wu S, Meltzer S, Cheng T, Jan LY, Jan YN, Cheng Y. Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature. 2017 Jul 6;547(7661):118-122. doi: 10.1038/nature22981. Epub 2017 Jun 26. PMID:28658211 doi:http://dx.doi.org/10.1038/nature22981
- ↑ Mascarenhas NM, Gosavi S. Understanding protein domain-swapping using structure-based models of protein folding. Prog Biophys Mol Biol. 2017 Sep;128:113-120. PMID:27867057 doi:10.1016/j.pbiomolbio.2016.09.013
- ↑ InterPro (IPR002110). Ankyrin repeat. Available at: https://www.ebi.ac.uk/interpro/entry/InterPro/IPR002110/. Accessed on 2025 June 19.
- ↑ Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006 Dec 26;45(51):15168-78. PMID:17176038 doi:10.1021/bi062188q
- ↑ Wang Y, Guo Y, Li G, Liu C, Wang L, Zhang A, Yan Z, Song C. The push-to-open mechanism of the tethered mechanosensitive ion channel NompC. Elife. 2021 Jun 8;10:e58388. PMID:34101577 doi:10.7554/eLife.58388
- ↑ 7.0 7.1 7.2 Hehlert P, Effertz T, Gu RX, Nadrowski B, Geurten BRH, Beutner D, de Groot BL, Göpfert MC. NOMPC ion channel hinge forms a gating spring that initiates mechanosensation. Nat Neurosci. 2025 Feb;28(2):259-267. PMID:39762662 doi:10.1038/s41593-024-01849-3
- ↑ 8.0 8.1 Cheng LE, Song W, Looger LL, Jan LY, Jan YN. The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron. 2010 Aug 12;67(3):373-80. PMID:20696376 doi:10.1016/j.neuron.2010.07.004
- ↑ 9.0 9.1 Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, Jan YN. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature. 2013 Jan 10;493(7431):221-5. PMID:23222543 doi:10.1038/nature11685
- ↑ 10.0 10.1 Effertz T, Wiek R, Göpfert MC. NompC TRP channel is essential for Drosophila sound receptor function. Curr Biol. 2011 Apr 12;21(7):592-7. PMID:21458266 doi:10.1016/j.cub.2011.02.048
- ↑ Schüler A, Schmitz G, Reft A, Özbek S, Thurm U, Bornberg-Bauer E. The Rise and Fall of TRP-N, an Ancient Family of Mechanogated Ion Channels, in Metazoa. Genome Biol Evol. 2015 Jun 22;7(6):1713-27. PMID:26100409 doi:10.1093/gbe/evv091